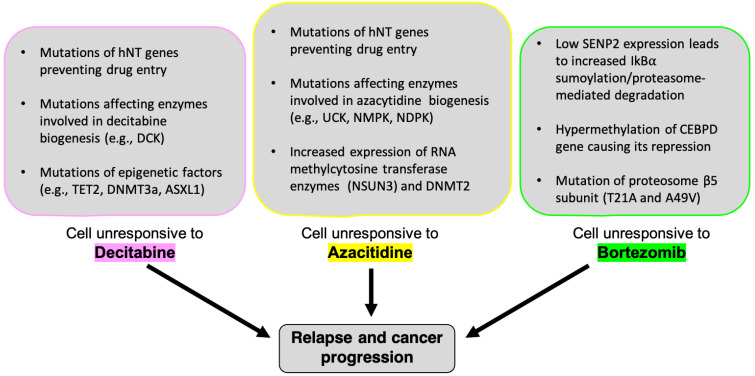
Figure 4.
Mechanisms of drug resistance in hematological malignancies. The mechanisms of cellular resistance to decitabine and azacitidine are still under investigation, however, several studies have begun to elucidate processes associated with resistance to both drugs. Mutations in epigenetic proteins (i.e., DNMT1, DNMT3a, TET2, and ASXL1) are often associated with decitabine [and azacitidine] resistance; these genetic alterations result in a hindered DNA methylation process and interfere with both drug’s ability to induce hypomethylation. Recent research has uncovered connections between azacitidine resistance and altered chromatin structures, specifically as it relates to RNA methyltransferases (RCMTs). In azacitidine responsive cells, RCMTs NSUN3 and DNMT2 bind specific RNA binding proteins and transcription factors to form an azacitidine-sensitive chromatin structure. However, drug resistant MDS, AML, and leukemic cells display high levels of NSUN1-BRD4-RNA-Pol2 binding; this binding complex forms an active chromatin structure that is insensitive to azacitidine. Additionally, mutations in enzymes involved in the biogenesis of decitabine (i.e., deoxycytidine kinase [DCK], nucleoside monophosphate kinase [NMPK]) and azacitidine (i.e., nucleoside monophosphate kinase [NMPK], uridine cytidine kinase [UCK]) contribute to cellular resistances to these drugs, alongside mutations in pyrimidine metabolic pathways which alter cancer cells’ ability to effectively process both drugs. Finally, nucleoside uptake transporter mutations contribute to such resistances, as these mutations prevent decitabine/azacitidine entry and metabolic activation in malignant cells. Bortezomib resistances involve a variety of genetic alterations and cellular pathways. Mutations in and/or overexpression of the β5 subunit gene of the proteasome’s 20S catalytic core are known to be involved in bortezomib resistances. More specifically, two specific amino acid swaps (i.e., T21A and A49V) alter the binding pocket of the β5 subunit, decreasing bortezomib’s affinity for the subunit’s active site. Given its activation and epigenetic effects in myeloma cells treated with bortezomib, CCAAT/enhancer binding protein delta (CEBPD) may also have a role in conferring bortezomib resistant to malignant cells. Hypermethylation of the CEBPD gene is characteristic in many patients suffering from AML. CEBPD is also highly expressed in cells treated with drugs [like bortezomib] that activate the p38 MAPK pathway, suggesting a synergistic interaction between this pathway and bortezomib that results in CEBPD activation. Additionally, bortezomib resistant cells display low levels of SENP2, a serine protease involved in the sumoylation of IkBα, and subsequent activation of NF-kB. With a wide array of [interlocking] mechanisms associated with drug resistance, future research should focus on deconstructing these pathways’ involvement in cancer prognosis; doing so will help develop targeted treatment regimens that contribute to long-lasting remission and patient survival.