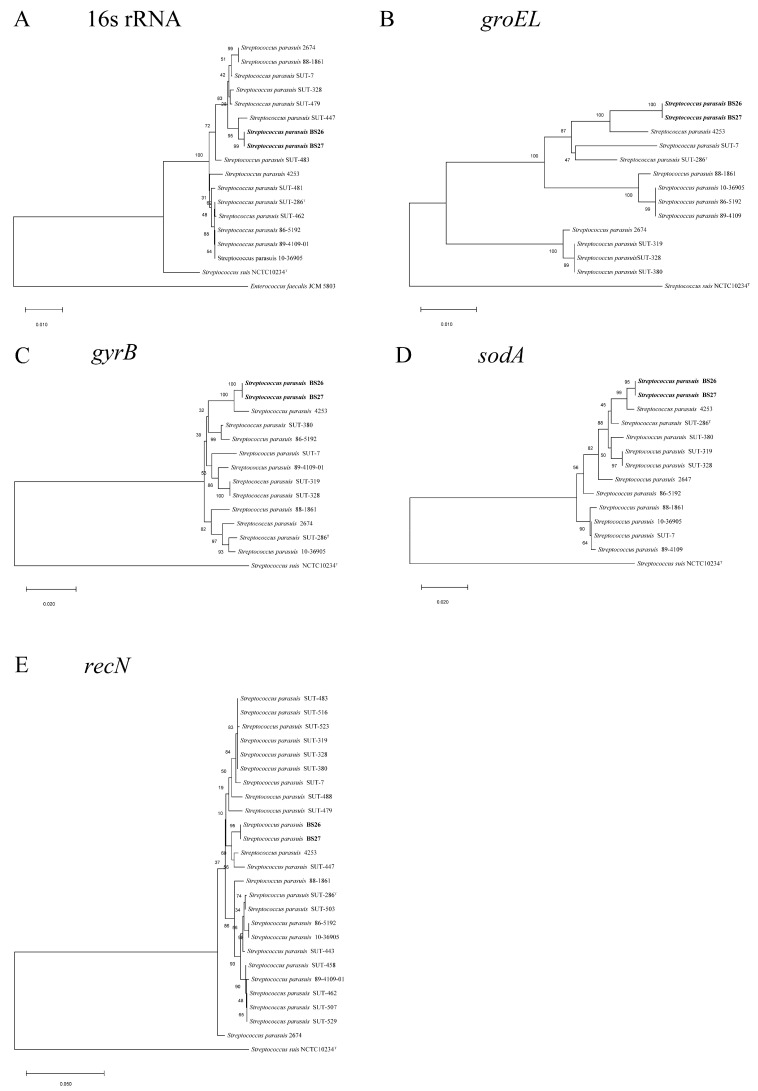
Figure 1.
Phylogenetic relationships of strains used in the present study. The trees were based on an alignment of full length of the 16S rRNA (A), groEL (B), gyrB (C), sodA (D), and recN (E) by using the neighbor-joining method. The corresponding sequence of E. faecalis JCM 5803 (for 16S rRNA gene) or S. suis type strain NCTC10234T (for groEL, gyrB, sodA, and recN genes) was used as an out-group to root the trees. The confidence values were obtained from 1000 replications. The bar represents sequence dissimilarity.