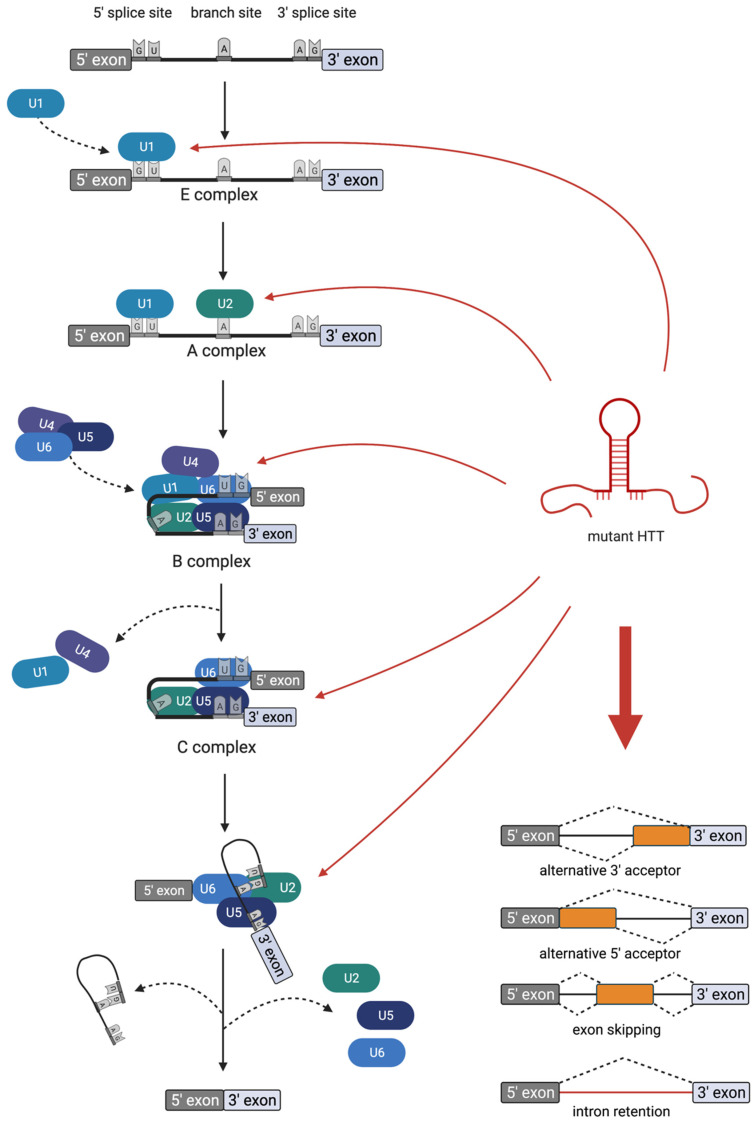
Figure 3.
Proteins that aberrantly bind to HTT RNA with expanded CAG repeats regulate splicing. A schematic of the splicing process is shown. RNA splicing is a process that removes non-coding sequences (introns) from pre-mRNA and joins the protein-coding sequences (exons) together, which is carried out by the spliceosome. The spliceosome consists of several proteins and small nuclear RNAs. The major spliceosome includes the small nuclear ribonucleoproteins (snRNPs) U1 (blue), U2 (green), U4 (violet), U5 (dark blue) and U6 (blue). These snRNPs sequentially attach to the pre-mRNA and carry out splicing, starting with U1, which binds at the 5′ splice site of the exon. U2 assembles at the branch site and then the U5 and U4–U6 complexes attach, resulting in the for-mation of the precatalytic spliceosome. Afterward, U4 and U1 are released from the pre-spliceosomal complex, the 5′ splice site gets cleaved and a looped lariat structure containing the intronic sequence is removed. Finally, the two exons are linked. HTT RNA with an expanded CAG repeat (hairpin depicted in red) aberrantly sticks to the snRNPs U1, U2, U4, U5 and U6. This results in aberrant splicing events, including intron retention, exon skipping or the use of alternative 3′ or 5′ acceptor sites. Created with BioRender.com (accessed on 22 March 2021).