Figure 1.
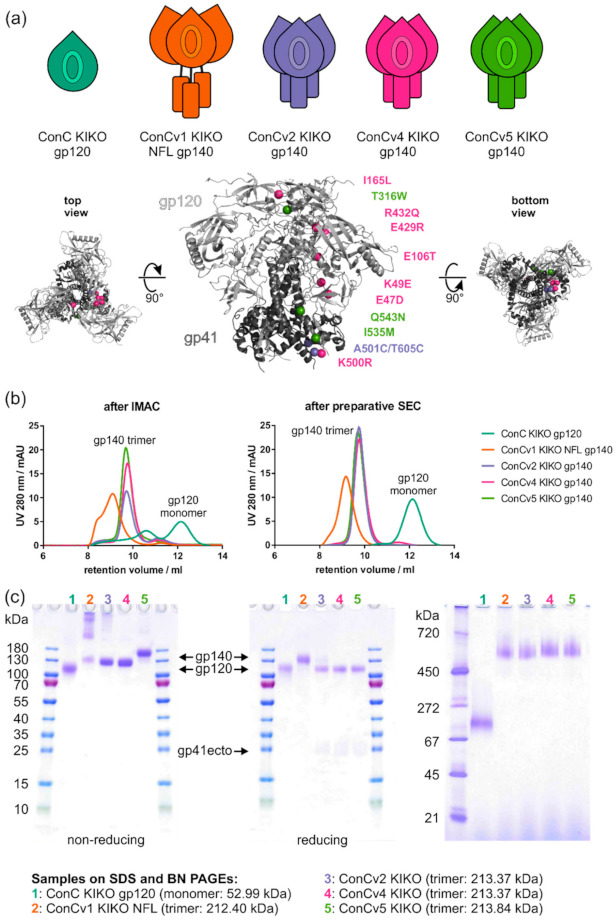
Purification of ConC Env variants for biophysical analyses and use as vaccines. (a) Schematic representation of the set of five ConC-derived proteins consisting of one gp120 monomer (ConC KIKO gp120, teal) and four gp140 trimers with stepwise increasing number of stabilizing modifications (ConCv1 KIKO NFL, orange; ConCv2 KIKO, lilac; ConCv4 KIKO, pink; ConCv5 KIKO, green). All included modifications are depicted and mapped on a pre-fusion Env trimer structure (PDB: 6CK9; ConC_Base0), as far as the respective regions were resolved. I559P first included in ConCv2 KIKO gp140; V65K first included in ConCv4 KIKO gp140; and H66R, A73C, and A561C first included in ConCv5 KIKO are hence not displayed. Colors are used according to the ConC Env version where the respective modification was first introduced. (b) Chromatograms of analytical size exclusion (SEC) runs with 10 µg of protein loaded directly after immobilized metal affinity chromatography (IMAC) (left) or after subsequent preparative SEC (right). (c) Non-reducing (left) and reducing (middle) SDS PAGEs for confirmation of correct cleavage and BN PAGE (right) for confirmation of purity of trimer in the final protein pool. Molecular weight for the gp120 monomer and the gp140 trimers are indicated, respectively. Two micrograms of protein were loaded per lane.
