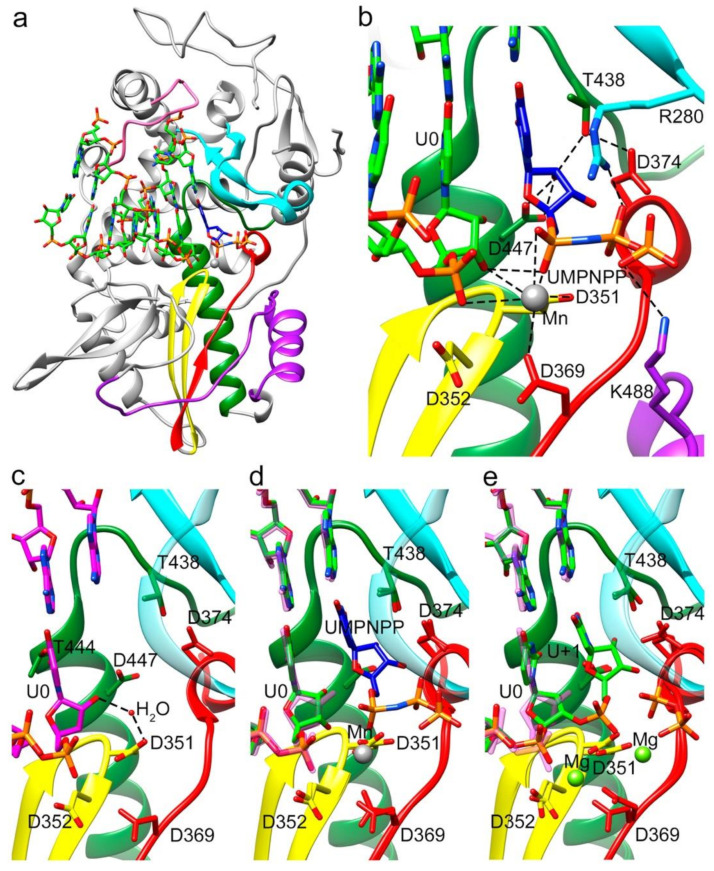
Figure 4.
Overall structure of the TaV RdRP in complex with RNA and the incoming nucleoside analogue uridine-5’-[(α,β)-imido]triphosphate (UMPNPP). (a) Side view of the overall structure of the Δ10-TaVpolΔ607-624/5′CAAAAUUU3′/UMPNPP complex enzyme (grey) with the conserved motifs and the bound RNA template/primer colored as in Figure 1. The substrate analogue is shown as sticks, colored according to atom type (C blue). (b) Close up view of the interactions involved in substrate analogue recognition. The side chains of residues D374 (motif A) and T438 and D447 (motif B) that participate in UMPNPP 2′-OH recognition are shown as sticks and explicitly labeled. One Mn2+ ion was found in the active site interacting with D351 (motif C), D369 (motif A) and the phosphate 1 moiety of the substrate. (c) Close up view of the active site region (same orientation as in B), in the structure of the TaVpolΔ607-624/5′CAAAAUUU3′ complex obtained in the absence of incoming nucleotides. The polymerase molecule is shown as green ribbons with the key active site chain depicted as sticks. (d) Superimpositions of the TaVpolΔ607-624/5′CAAAAUUU3′ complex structures in the absence (RNA carbon atoms in magenta) and in the presence of the UMPNPP analogue (RNA carbon atoms in green; UMPNPP in blue). The Mn2+ ion is shown as a grey sphere. (e) Superimposition of the TaVpolΔ607-624/5′CAAAAUUU3′ complex structures in the absence of the incoming substrate (RNA carbon atoms in magenta) and after UMP incorporation, trapped before translocation (RNA carbon atoms in green). The two bound Mg2+ ions are shown as green spheres.