Figure 1.
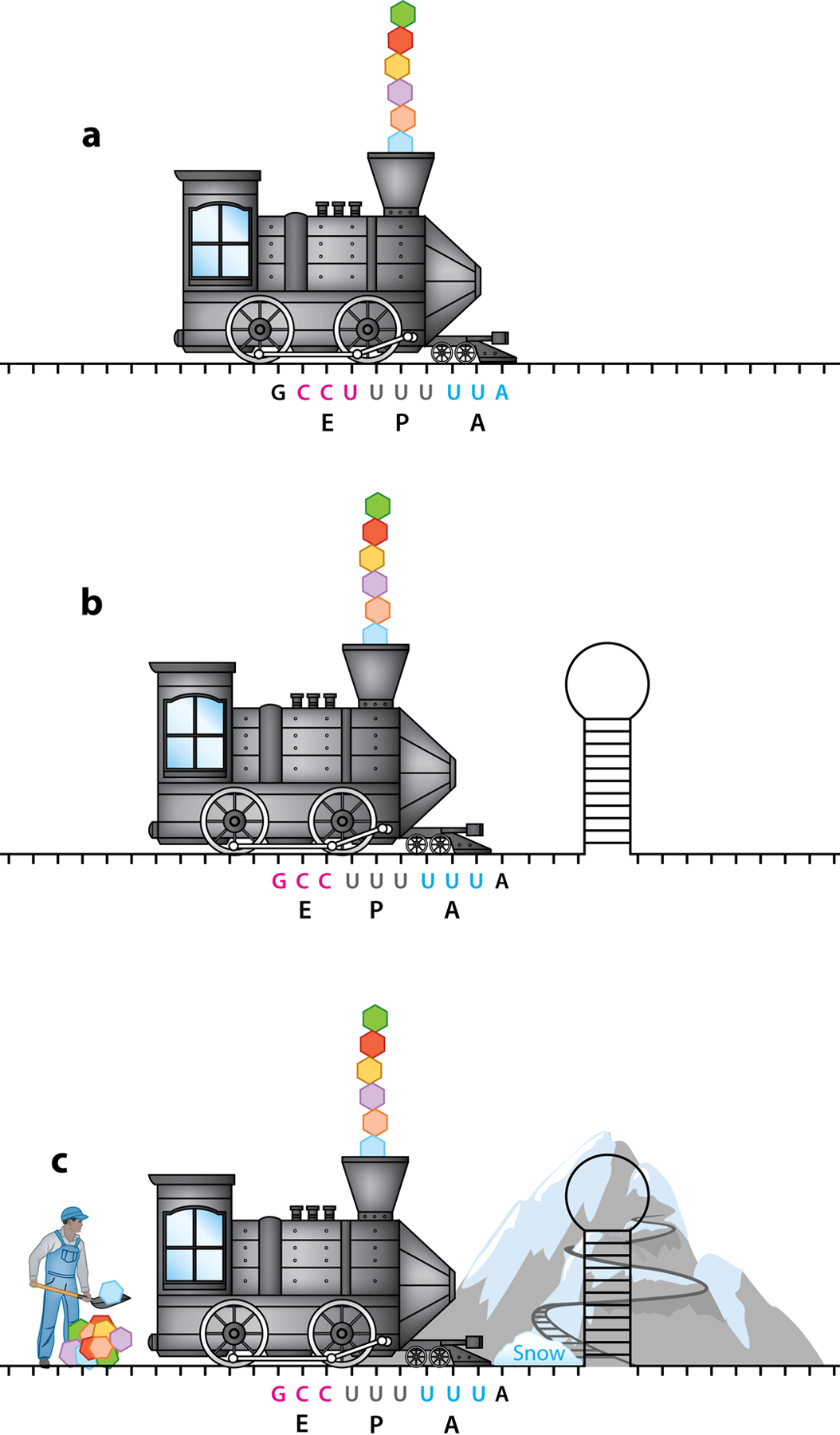
Schematic of how cis and trans elements affect viral and host PRF translation. The train represents the ribosome, and the tracks represent the mRNA. The slippery-site is U UUU UUA with the designated codons in the E, P, and A sites shown in pink, gray, and blue, respectively. The newly formed polyprotein chain is being released from the train’s smokestack. (a) The ribosome chugs along the mRNA, translating a new polyprotein. The E, P, and A sites are labeled along with the corresponding codons. (b) PRF is usually stimulated by an mRNA secondary structure downstream of the slip-site. An encounter with the mRNA stem-loop allowed the train to slip back 1 nucleotide (−1 PRF; shown here), and now new codons are in the EPA sites of the ribosomes. (c) cis and trans regulatory factors have been identified to modulate PRF. These include (but are not limited to) the mountain, which represents proteins or microRNA that bind and stabilize the mRNA stem-loop and forces the train to slow down, enhancing PRF; the snow, which is a host or viral protein that causes the ribosome train to pause, which is quite prevalent; and the concentration of translational factors including tRNAs (depicted by the worker shoveling tRNAs into the train) that influence kinetics and thermodynamics of translation, which is prevalent in +1 PRF. Abbreviations: EPA, exit, peptidyl, and aminoacyl; mRNA, messenger RNA; PRF, programmed ribosomal frameshifting; tRNA, transfer RNA.
