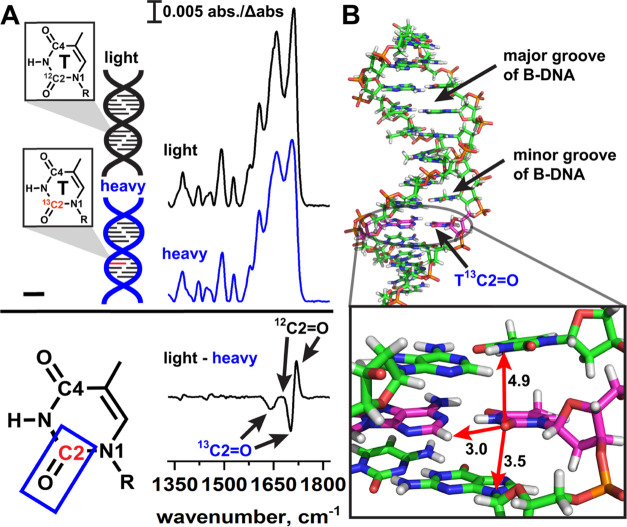
Figure 2.
Isotope editing and spectral subtraction isolate isotope signals from individual T bases in DNA duplexes. (A) In isotope editing experiments, an isotopically labeled base is site-specifically incorporated into a DNA duplex (heavy, blue) and the heavy IR spectrum is subtracted from the IR spectrum of a light duplex (black), which has an identical sequence. In the “light–heavy” subtraction spectrum, signals due to the light 12C=O appear as positive bands, and signals due to the heavy 13C=O appear as negative bands. Signals that arise from vibrations in which the C2 atom does not participate are canceled out, thus reducing spectral congestion so that C=O stretching of a specific bond of an individual DNA base can be observed in the duplex environment. (B) B-form DNA duplex illustrating the local environment of a TC2=O bond. Arrows indicate nearby functional groups on neighboring bases that may impact the stretching frequency. Distances are in angstrom.