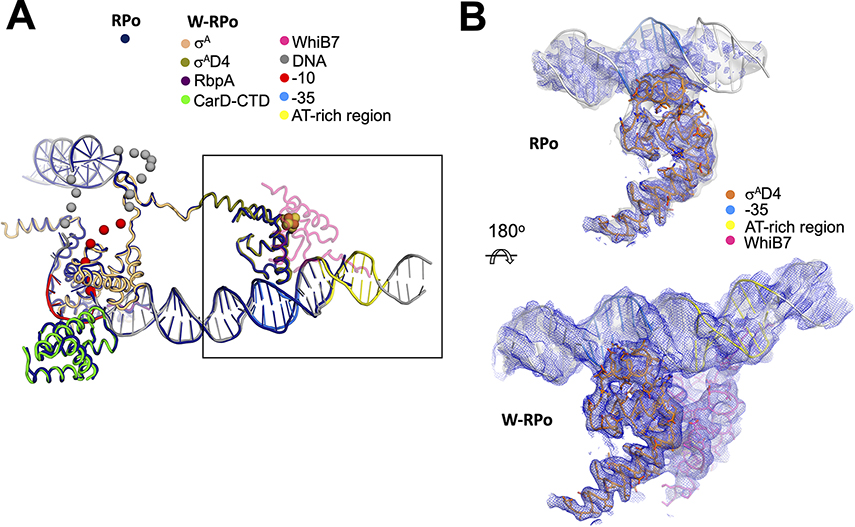
Figure 3. WhiB7 stabilizes σAD4 contacts with the −35 promoter element.
A. The W-RPo and RPo structures were superimposed by aligning α-carbon positions of σA, the CarD-CTD, and the RbpA-linker (root-mean-square deviation of 0.535 Å over 417 α-carbon atoms). The resulting DNA structures closely superimposed. WhiB7 is shown in faded pink to increase the clarity of σAD4. The disordered and unmodeled regions of the transcription bubble are indicated by spheres.
B. The sharpened (blue mesh) and local-resolution filtered (gray transparent surface) cryo-EM maps are show for RPo (top) and W-RPo (bottom), with the structures superimposed. The view shows a zoom-in of the boxed area in (A), reoriented as shown. Cryo-EM density for σAD4 contacts with the −35 promoter region in RPo is weak in the sharpened map, and density in the local-resolution filtered map disappears about six base-pairs upstream of the −35 motif. Cryo-EM density for the σAD4 contacts with the −35 promoter region in W-RPo is well-resolved and better connected in both the sharpened (blue mesh) and local-resolution filtered (light grey surface) maps, and density extends well upstream of the AT-rich DNA bound by the WhiB7 AT-hook. Maps were normalized in PyMOL and contoured at 4σ.