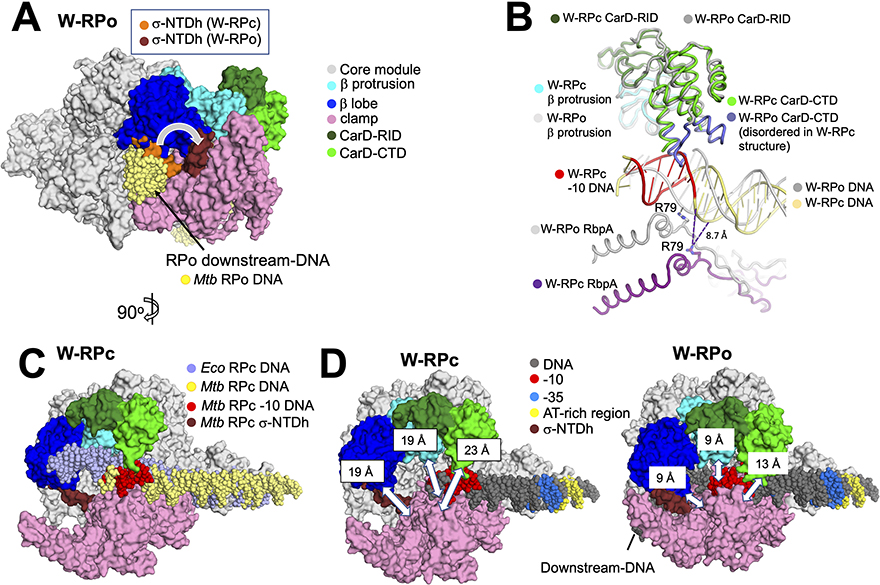
Figure 5. Comparisons of structural differences between W-RPc, W-RPo and Eco RPc.
A. Structural alignment of the core module (Boyaci et al., 2018) of W-RPc to W-RPo shows that in WRPc, σA N-terminal domain helix (W-RPc σ-NTDh; orange) occupies the space in the channel where the downstream-DNA (colored yellow in this model) eventually rests in W-RPo. Upon W-RPo formation, the helix is moved (W-RPo σ-NTDh; brick red) to interact with the β- lobe so that the DNA can be placed in its RPo position. The structure shown is W-RPo with the superimposed W-RPc σ-NTD. Core, β lobe, β protrusion, clamp and CarD are colored as listed in key on the right.
B. Structural alignment of the β-protrusion between W-RPo and W-RPc reveals that CarD from W-RPc would clash with the double-stranded −10 element DNA. In W-RPc, the loops on the CarD-CTD (colored purple) are disordered, presumably due to the clash with the double-stranded DNA. In W-RPo, RbpA-R79 is positioned to form an ionic bond (2.0 Å distance) with OP1 of the non-template strand DNA −14 position, an interaction shown previously to be critical for RbpA transcription activation function (Hubin et al., 2017). However, RpbA-R79 in W-RPc is 19 Å away from the non-template DNA −14 phosphate oxygens, and the closest nucleotide is the non-template strand −16 position, located 8.7 Å away. Thus, RbpA-DNA backbone contacts are not established in RPc. Dashed lines indicate distances, with the distance of 8.7 Å noted for reference.
C. Structural alignment of the core module of Eco RPc (Boyaci et al., 2018) and W-RPc reveals that the downstream DNA trajectory changes due to CarD-CTD (green). The structure shown is W-RPc, except for the Eco RPc DNA (slate). Core, β lobe, β protrusion, clamp and CarD are colored as listed in key in (A).
D. Distances between the clamp and the β-lobe (α-carbons of σA-F233 and β-G284), the clamp and β-protrusion (α-carbons of σA-R309 and b-E402), and the clamp and CarD-CTD (a-carbons of σA-T345 and CarD-R87) were measured for W-RPc (left) and W-RPo (right) using PyMOL (The PyMOL Molecular Graphics System, Version 2.3.5 Schrodinger, LLC). These distances are illustrated by white double-edged arrows and noted on the “top” pincer domains. Core, β lobe, β protrusion, clamp, CarD, σ-NTDh and DNA are colored as listed in key in (A).