Figure 1:
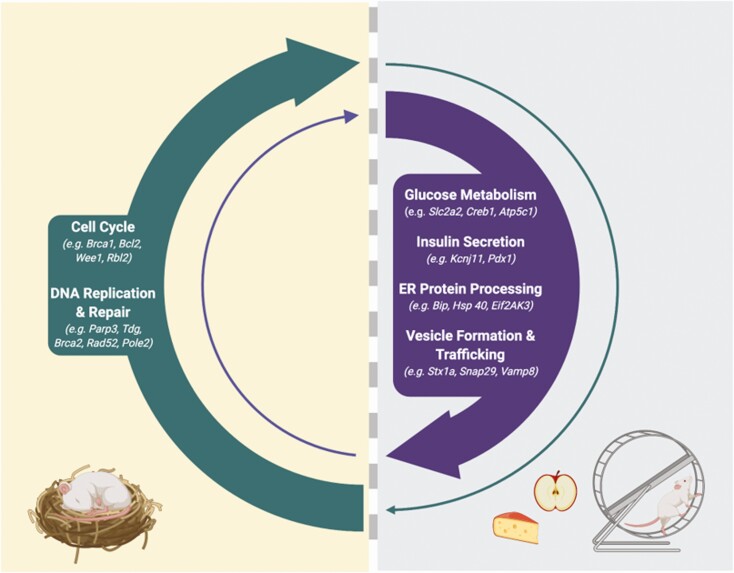
Temporal transcriptional profiling of rodent islets suggests that the circadian clock transcriptionally gates important and diverse biological pathways. During the active phase, when food is anticipated, the circadian clock drives the expression of key insulin secretory genes, including genes involved in glucose metabolism (eg, Slc2a2 (9, 30), Gnaq (9), Creb1 (9), Creb3 (9), Cacna1c (9), Atp5c1 (30), Cox4L1 (30), Ndufa2 (30)), insulin secretion (Slc2a2 (9, 30), Creb3L1 (30), Pdx1 (30), KcnJ11 (30)) ER function and protein processing (eg, Bip, Hsp40, Hsp70, Grp94, Erolβ, Eif2Ak3 (Perk), Dsk2) (30), and vesicle maturation, trafficking and exocytosis (eg, Stx1a (9), Vamp8 (9), Vamp4 (30), Bet1 (30), Snap29 (30)). During the inactive phase of the day, these pathways are suppressed, and pathways involved in growth and repair, including cell cycle control genes (eg, Bcl2, Brca1, Ccnb2, Cdk6, Cdc20, Wee1, Cdkn2b, Rbl2, Hus1, Gadd45a) (27), and DNA replication and repair (eg, Parp3, Tdg, Brca2, Rad52, Pole2) (30) are activated. From these transcriptional programs, we speculate that the beta cell circadian oscillator drives periods of insulin secretion “priming” when food is anticipated with alternating periods of rest and recovery. This hypothesis is based on rodent studies and notably, rodents are nocturnal in nature. Similar temporal transcriptional profiling studies in human islets are currently lacking but are needed to understand how the circadian clock may transcriptionally gate such pathways in diurnal humans. Created with BioRender.com.