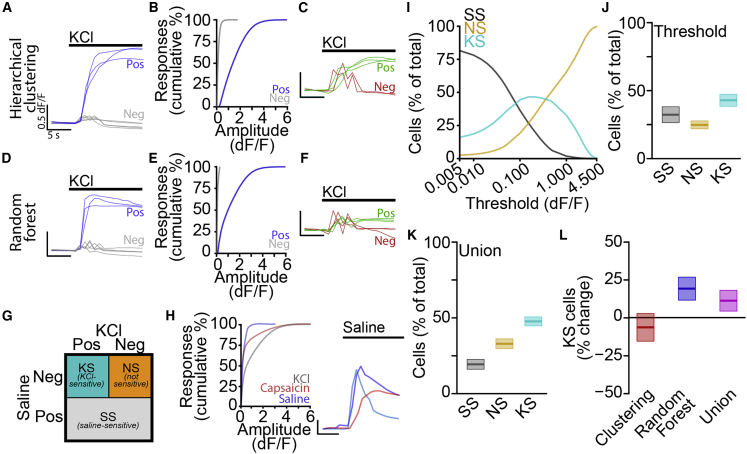
Figure 2.
Machine learning-enabled classification of automated calcium imaging responses
(A) Traces of KCl-dependent responses classified as positive (blue) and negative (gray) by hierarchical clustering. Scale bars, 5 s, 0.5 dF/F.
(B) Cumulative distributions of positive (blue) and negative (gray) response amplitudes.
(C) Traces of small amplitude KCl-dependent responses classified as negative (red) and positive (green) by hierarchical clustering. Scale bars, 5 s, 0.5 dF/F.
(D) Traces of KCl-dependent responses classified as positive (blue) and negative (gray) by random forest model. Scale bars, 5 s, 0.5 dF/F.
(E) Cumulative distributions of positive (blue) and negative (gray) response amplitudes.
(F) Traces of small amplitude KCl-dependent responses classified as negative (red) and positive (green) by random forest model. Scale bars, 5 s, 0.5 dF/F.
(G) Cell classification scheme.
(H) Cumulative distributions of response amplitudes (left) for saline (blue), capsaicin (red), and KCl (gray) with traces (right, colors indicate individual cells). Scale bars, 5 s, 0.5 dF/F.
(I) Percentage of total cells classified as saline sensitive (SS) (gray), not sensitive (NS) (orange), and KCl-sensitive (KS) (cyan).
(J) Percentage of cells classified as SS (gray), NS (orange), and KS (cyan) by using standard threshold of 0.15 dF/F as mean ± SEM.
(K) Percentage of cells classified as SS (gray), NS (orange), and KS (cyan) by using the union of hierarchical clustering and random forest classifiers mean ± SEM.
(L) Percent change from threshold-based analysis in percentage of KS cells by using hierarchical clustering (red) or random forest (blue) classifiers or the union (magenta) as mean ± SEM.