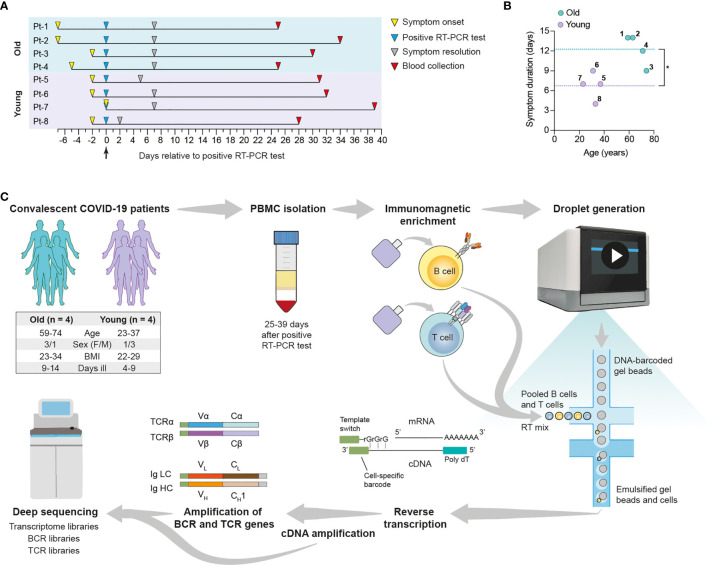
Figure 1.
Overview of single-cell transcriptome and immune receptor profiling of convalescent COVID-19 patient lymphocytes. Convalescent COVID-19 patients enrolled in the SERO-BL-COVID-19 study were selected according to their age for single-cell sequencing analysis of their T cells and B cells. (A) Timeline illustrates symptom onset, symptom resolution and collection of blood samples from individual patients relative to the time of positive SARS-CoV-2 RT-PCR test (day 0). (B) Graph displays the ages and duration of COVID-19 symptoms in individual patients. Dotted lines show the mean duration of symptoms in the young (y = 6.75 days) and old (y = 12.25 days) groups. A significant difference in symptom duration between groups is indicated with an asterisk (p = 0.0127; unpaired t-test). (C) Single-cell sequencing protocol. Whole blood was collected following the resolution of COVID-19 symptoms and subjected to density gradient separation for isolation of PBMC. T cells and B cells from individual patients were purified from PBMC using negative immunomagnetic enrichment, pooled (intra-patient) and prepared for droplet generation using the 10x Genomics Chromium system. Single cells were emulsified with DNA-barcoded gel beads and mRNA transcripts were reverse-transcribed within droplets, resulting in the generation of first-strand cDNA molecules labelled with cell-specific barcodes at their 3’ ends (added by template switching). Emulsions were disrupted and cDNA was amplified by means of PCR for further processing of transcriptome libraries. Transcriptome libraries from individual patients were indexed and multiplexed for deep sequencing using the Illumina NovaSeq platform. Targeted enrichment of recombined V(D)J transcripts was performed by PCR and the resulting products were processed for the generation of BCR and TCR libraries, which were then indexed, multiplexed and deep-sequenced.