FIGURE 1.
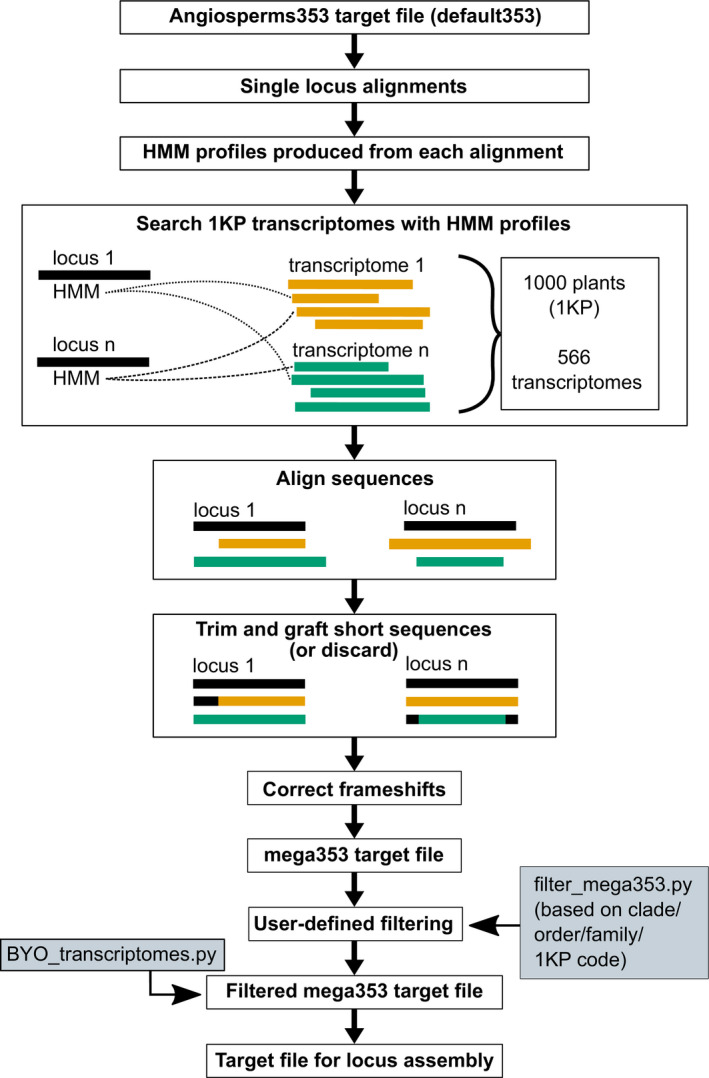
Overview of the steps involved in creating the mega353 target file. First, loci in the default353 file are aligned and hidden Markov model (HMM) profiles are created for each locus. The HMM profiles are used to identify these loci in the 1KP transcriptomes, and transcript hits are added to the alignment. The alignment of each locus is then trimmed, grafted if necessary, a frameshift correction is performed, and all loci are combined in the mega353 target file. The gray boxes indicate steps the user can take to modify the mega353 target file. The mega353 target file can be filtered (based on sample identifiers in the filtering_options.csv file) to select samples included in the target file. The BYO_transcriptome.py script can be used to add GenBank or personal transcriptomes to the filtered mega353 target file.
