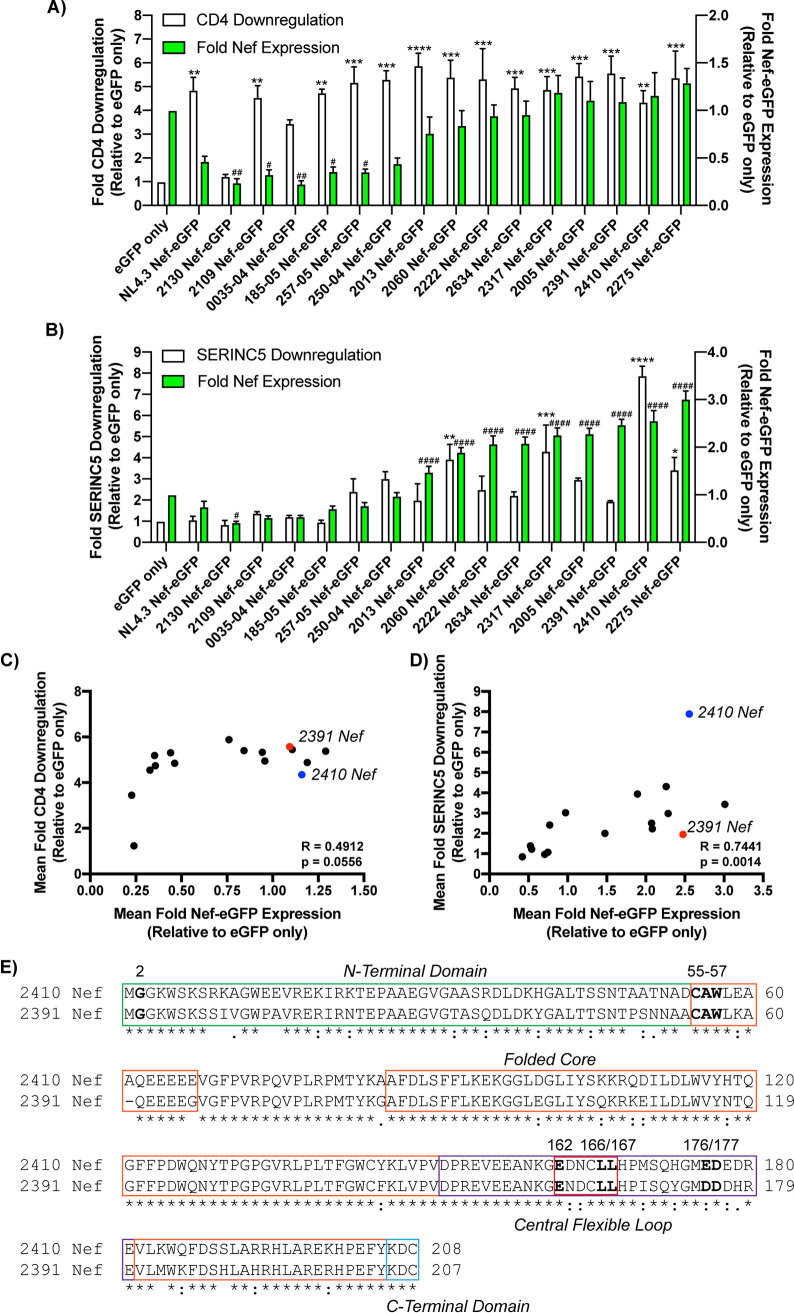
FIG 1.
Primary Nef isolates displayed diverging Nef functionality. (A) CD4+ HeLa cells expressing Nef isolates were stained for cell surface CD4 and analyzed by flow cytometry. Shown is a summary of the fold CD4 downregulation ability (±SE) on the left y axis and fold Nef expression (±SE) on the right y axis for each Nef isolate (n = 3). (B) CD4+ HeLa cells coexpressing each Nef isolate and SERINC5.intHA were stained for cell surface SERINC5 and analyzed by flow cytometry. Shown is a summary of the fold SERINC5 downregulation ability (±SE) on the left y axis, and fold Nef-expression (±SE) on the right y axis for each Nef isolate (n = 3). Significance of Nef-eGFP expression and CD4/SERINC5 cell surface downregulation are denoted by # and ∗, respectively, upon comparison to the eGFP only control. Correlation analysis between the mean fold Nef-eGFP expression (x axis) and mean fold CD4 downregulation (C) and mean fold SERINC5 downregulation (y axis) (D) are shown for the 15 described primary Nef isolates and NL4.3 Nef. The blue and red dots indicate the 2410 Nef-eGFP and 2391 Nef-eGFP samples, respectively. Statistical analysis was conducted using the Spearman rank correlation test. (E) Amino acid sequence alignment of 2410 and 2391 Nef. Boldfaced Nef residues denote shared residues/motifs involved in Nef-mediated CD4 downregulation or enhancing virion infectivity in the presence of SERINC5. The location of the specific Nef domains, including the N-terminal domain (green), folded core (orange), central flexible loop (purple), Nef dileucine motif (red), and the C-terminal domain (blue), are highlighted in the appropriate boxes. An asterisk indicates positions with a fully conserved residue between the isolates. A colon indicates conservation of residues with similar properties (scoring >0.5 in the Gonnet PAM 250 matrix). A period indicates conservation of residues with less similar properties (scoring ≤0.5 and >0 in the Gonnet PAM 250 matrix). eGFP, enhanced green fluorescent protein; SE, standard error; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; #, P ≤ 0.05; ##, P ≤ 0.01; ####, P ≤ 0.0001.