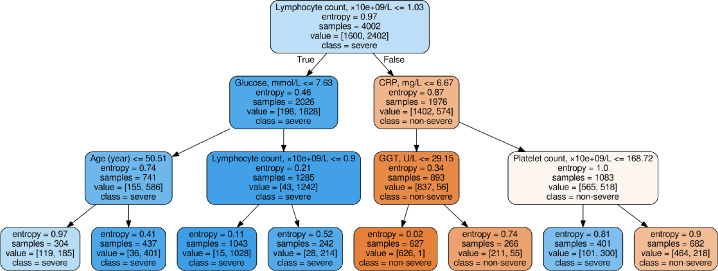
Fig 6. Results from the DT model trained on the biochemistry data.
The DT is a directed graph based model which consists of nodes, leaves and edges. Each node contains a decision with two output edges for True and False rule, with the exception of the terminal nodes, which are called leaves and denote the class label. For each sample we can manually move from the root to a leaf by checking each node’s rules. As this model is prone to overfitting, we limited the depth of the tree to 3 and the minimum number of samples in the leaf to 120 (5% of the larger class). This approach allows monitoring the most stable and important decision rules which distinguish severe and non-severe cases.