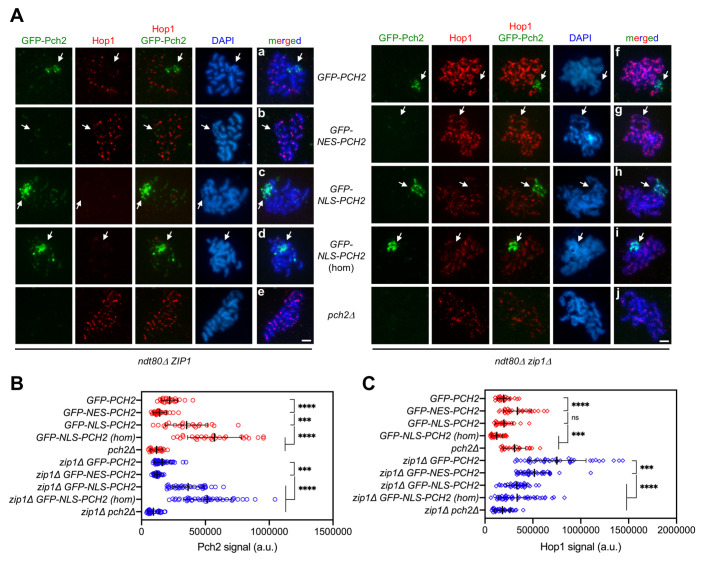
Fig 4. Impact of GFP-NES-Pch2 and GFP-NLS-Pch2 on the pattern of Hop1 chromosomal localization.
(A) Immunofluorescence of meiotic chromosomes stained with anti-GFP antibodies (to detect GFP-Pch2; green), anti-Hop1 antibodies (red) and DAPI (blue). Representative ZIP1 and zip1Δ nuclei, as indicated, are shown. Arrows point to the rDNA region. Spreads were prepared from ndt80Δ strains at 24 h. Scale bar, 2 μm. (B, C) Quantification of the GFP-Pch2 and Hop1 signal, respectively. Error bars: SD; a.u., arbitrary units. Red and blue symbols correspond to ZIP1 and zip1Δ strains, respectively. Strains are: DP1654 (GFP-PCH2), DP1725 (GFP-NES-PCH2), DP1729 (GFP-NLS-PCH2), DP1768 (GFP-NLS-PCH2 [hom]), DP1058 (pch2Δ), DP1655 (zip1Δ GFP-PCH2), DP1726 (zip1Δ GFP-NES-PCH2), DP1730 (zip1Δ GFP-NLS-PCH2), DP1769 (zip1Δ GFP-NLS-PCH2 [hom]) and DP881 (zip1Δ pch2Δ).