Summary
The plant cell wall provides mechanical strength to support plant growth and development and to determine plant architecture. Cellulose and mixed‐linkage glucan (MLG) present in primary cell wall, whereas cellulose, lignin and hemicellulose exist in secondary cell wall. Biosynthesis of the cell wall biopolymers needs the coordinated transcriptional regulation of all the biosynthetic genes. The module of OsmiR166b‐OsHox32 regulates expression levels of the genes related to biosynthesis of MLG, cellulose and lignin. Transgenic plants knocking down miR166b (STTM166b) by short tandem target mimic (STTM) technology or overexpressing OsHox32 (OEHox32) showed drooping leaves and brittle culms. Due to accumulation of less lignin and cellulose, the cell wall thickness of STTM166b and OEHox32 plants was reduced when compared to that of wild‐type plants. Overexpression of miR166b (OE166b) in rice plants or knocking down of OsHox32 by RNA interference (RNAiHox32) led to increased thickness of cell walls and enhanced mechanical strength of culms. Molecular analyses showed that OsmiR166b‐OsHox32 pair regulates cell wall‐related gene expression. OsHox32 binds to the promoters of OsCAD2 and OsCESA7 to suppress the expression levels of these two genes. The suppression of OsCAD2 is synergistic when OsHox32 is co‐expressed with OSH15 (Oryza sativa homeobox 15). OsHox32 interacts with OSH15, and the START domain of OsHox32, harbouring the miR166b cleavage site, is required for the interaction of these two proteins. Our results demonstrate that OsmiR166b‐OsHox32 pair plays important roles not only in plant growth and development but also in plant architecture by regulating the cell wall‐related gene expression.
Keywords: cell wall, cellulose, Hox32, lignin, mechanical strength, miR166b, rice
Introduction
The plant cell wall is a complex and heterogeneous matrix of polysaccharides, glycoproteins, solutes and enzymes, which governs the morphology, growth and development of plants. Primary cell wall is a polymeric network of crystalline cellulose microfibrils, pectic polysaccharides and xyloglucans, which possesses a remarkable combination of strength and pliancy that enable cells to withstand mechanical forces and to create space for growth (Cosgrove and Jarvis, 2012). Secondary cell wall forms a thick layer rich in cellulose, hemicellulose and lignin to provide mechanical strength and rigidity allowing vascular plants reach great heights and compete for light (Taylor‐Teeples et al., 2015). Synthesis of these cell wall‐related components requires expression of sets of specific genes that are regulated by multiple tiers of transcription factors. Cellulose is synthesized by rosette complexes that consist of multiple cellulose synthase (CESA) proteins. In rice, CESA1, 3 and 8 and CESA4, 7 and 9 are required for cellulose biosynthesis during primary and secondary cell wall formation, respectively (Tanaka et al., 2003; Wang et al., 2010). Lignin biosynthesis is controlled by a set of genes. For example, PAL1 (phenylalanine ammonia lyase 1), C4H (cinnamate 4‐hydroxylase) and 4CL3 (4‐hydroxycinnamate: CoA ligase 3) are responsible for the first three steps of lignin biosynthesis, while COMT (caffeic acid 3‐O‐methyltransferase) and CAD (cinnamyl alcohol dehydrogenase) catalyse the last two steps of biosynthesis (Zhao and Dixon, 2011). Plant growth and development need biosynthesis of the cell wall components that are controlled by different sets of biosynthetic genes, which are in turn controlled by the master switches of NAC domain and homeodomain‐leucine zipper Class III (HD‐ZIP III) transcription factors (Bhargava et al., 2010; Kubo et al., 2005).
Leaf architecture, including leaf shape and leaf angle, is an important agronomic trait that contributes largely to yield by affecting photosynthesis rate and planting density. Moderate width and erectness of leaves make rice plants have an ideal canopy which is the long sought target in rice breeding (Peng et al., 1999). As the importance of rice leaf architecture, scientists have paid much attention to the molecular mechanisms of leaf shape and leaf angle formation, and thus, many genes related to leaf architecture have been identified. The NAC domain transcription factors, such as VASCULAR‐RELATED NAC‐DOMAIN 7 (VND7), SECONDARY WALL‐ASSOCIATED NAC DOMAIN PROTEIN 1 (SND1) and NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1 (NST1), act as master regulators of the secondary cell wall biosynthesis (Endo et al., 2015). The Arabidopsis snd1 nst1 double‐mutant showed pendent inflorescence stem and reduced stem strength due to the defective deposition of cellulose and lignin (Zhong et al., 2011). Rice transcription factors SECONDARY WALL NAC DOMAIN PROTEINs (OsSWNs) regulate secondary cell wall formation, and repression of OsSWNs expression in rice resulted in drooping leaves and reduced plant height (Yoshida et al., 2013).
Culm stiffness is related to lodging resistance that plays important roles in grain yield and quality. Several factors such as the thickness and the chemical contents of cell walls have been reported to affect culm stiffness. Different genetic approaches have been applied to improve rice lodging, for example, the semidwarf gene sd‐1 has been used to reduce plant height and relatively increase sturdiness of the basal culms leading to the improvement of lodging (Sasaki et al., 2002). In general, lignin and/or cellulose determine physical strength of culms, thus reduced contents of lignin and/or cellulose lead to brittle culms (Li et al., 2003; Vega‐Sánchez et al., 2012). The maize brown midrib 1 (bm1) mutant, harbouring a mutation in the lignin biosynthetic CAD gene, shows a substantial reduction in the total lignin content (Halpin et al., 1998). The rice flexible culm 1 (fc1) mutant was identified as disrupting the CAD7 gene by a T‐DNA insertion, and the mutation caused a reduction in cell wall thickness and a decrease in lignin content, leading to reduced mechanical rigidity of culms (Li et al., 2009). Rice GOLD HULL AND INTERNODE 2 (GH2) gene encodes CAD2, and the gh2 mutant is lignin‐deficient and shows reduced lignin content in cell walls (Zhang et al., 2006).
The Arabidopsis KNOX protein BP (BREVIPEDICELLUS) regulates lignin content by suppressing the expression of a set of lignin biosynthetic genes, including At4CL1 (4‐coumarate: CoA ligase 1), AtC4H, PAL1, CCoAOMT (caffeoyl‐CoA O‐methyltransferase) and COMT1. EMSA (Electrophoretic mobility shift assay) demonstrated that BP binds to the promoters of COMT1 and CCoAOMT and suppresses the expression of these two genes (Mele et al., 2003). A rice KNOX gene Oryza sativa homeobox 15 (OSH15) is highly expressed in the abscission zone and OSH15 protein suppresses the expression of CAD2 by binding to its promoter, which leads to reduced lignin accumulation in the abscission zone and consequentially causes grain shattering (Yoon et al., 2017).
Homeodomain‐leucine zipper (HD‐ZIP) transcription factor family is unique to the plant kingdom and classified into four groups (I–IV) (Elhiti and Stasolla, 2009). It has been reported that expression of HD‐ZIP III genes is regulated by miR165/166 in Arabidopsis and by miR166 in rice at the posttranscriptional level (Rhoades et al., 2002; Zhang et al., 2018). Short Tandem Target Mimic (STTM) technology has been used to study the function of microRNAs (Gao et al., 2018; Yan et al., 2012). Our previous study showed that OsmiR166b is predicted from a QTL region contributing to grain yield of a heterotic rice hybrid (Fang et al., 2013; Li et al., 2000), and however, the function of OsmiR166b in rice growth and development is still elusive. In this study, we studied the function of OsmiR166b and its predicted target gene OsHox32, a member of the HD‐ZIP III group, in plant architecture formation. The OsmiR166b‐OsHox32 module affects leaf shape and culm rigidity by regulating expression levels of the cell wall‐related genes, leading to altered thickness of cell walls and mechanical strength of plants. The transcription factor OsHox32 suppresses the expression of OsCAD2 and OsCESA7 by binding to their respective promoter, thus leading to reduced lignin and cellulose contents.
Results
STTM166b and OEHox32 plants show drooping leaves and brittle culms
OsmiR166b has been shown to target genes encoding START domain‐containing proteins (http://structuralbiology.cau.edu.cn/PNRD/). To further study the function of OsmiR166b in rice, we selected OsHox32 (Loc_Os03g43930), one of the START domain‐containing genes, as the candidate target to pair with OsmiR166b (Figure S1a). To confirm that OsHox32 is an authentic target of OsmiR166b, we randomly mutated the nucleotides in the OsmiR166b‐cleaving site of OsHox32 and performed RT‐PCR analysis. The results confirmed that OsmiR166b cleaves OsHox32, but not the mutated mOsHox32 (Figure S1b). We generated transgenic plants either knocking down OsmiR166b (STTM166b) by STTM technology or overexpressing OsmiR166b (OE166b) by overexpressing OsMIR166b. Meanwhile, we also produced transgenic plants overexpressing or suppressing OsHox32. Expression levels of the manipulated genes in the homozygous lines were analysed by qRT‐PCR, and two representative lines for each gene transformation were investigated (Figure S2a, b).
When grown in the paddy field, STTM166b and OsHox32 overexpression (OEHox32) plants displayed similar phenotypes, whereas OE166b transgenic plants phenocopied OsHox32 suppression (RNAiHox32) plants (Figure 1a). Detailed investigation showed that transgenic plants displayed multiple phenotypic alterations in comparison to wild‐type plant ZH11. Although OE166b and RNAiHox32 transgenic plants did not show significant difference in plant height, STTM166b and OEHox32 showed reduced plant height when compared to wild type (WT) (Figure S2c, d). When compared the different internodes of transgenic plants, the first and the second internodes of STTM166b and OEHox32 were significantly shorter than those of ZH11 (Figure S3a, b). Leaves of STTM166b and OEHox32 were adaxially rolled and drooping although those of OE166b and RNAiHox32 did not show difference in comparison with the leaves of ZH11 (Figure 1b, Figure S3c, d). Another prominent feature of STTM166b and OEHox32 plants is the brittle culm that can be broken easily by bending (Figure 1c). Other agronomic traits of the transgenic plants were investigated and listed in Table S1. These phenotypic alterations imply multiple functions of OsmiR166b and OsHox32 in plant growth and development.
Figure 1.

Morphology of transgenic plants. (a) Morphology of wild type (ZH11) and OsmiR166b and OsHox32 transgenic plants at tillering stage. Bars = 10 cm. (b) Flag leaves from wild‐type and transgenic plants. (c) Breaking points of the culms from ZH11 and transgenic rice plants. Arrows indicate the breaking points.
OsmiR166b‐OsHox32 pair regulates cell wall thickness
Reduction in the mechanical strength of culms reflects changes in structure and/or composition of the cell walls. In rice plants, cells that are under the epidermis and around the peripheral vascular tissues provide mechanical support for plants. To investigate cell wall morphology, we used scanning electron microscopy (SEM) and transmission electron microscopy (TEM) to examine stem and leaf cells. SEM observation of the culm sections revealed that sclerenchyma cells of ZH11, OE166b and RNAiHox32 plants were almost completely filled up (Figure 2a–f), whereas those cells of STTM166b and OEHox32 culms contained cavities (Figure 2g–j). Furthermore, TEM observation of the leaf sections showed that cell walls of the sclerenchyma cells under epidermis of OE166b and RNAiHox32 plants were significantly thicker than those of ZH11 (Figure 3a–f), whereas cell walls of STTM166b and OEHox32 plants were significantly thinner than those of ZH11 (Figure 3g–j). Statistical data of the cell wall thickness were shown in Figure 3k. These findings suggest that the OsmiR166b‐OsHox32 pair plays important roles in modulating cell wall thickness, which determines the mechanical strength of plants.
Figure 2.
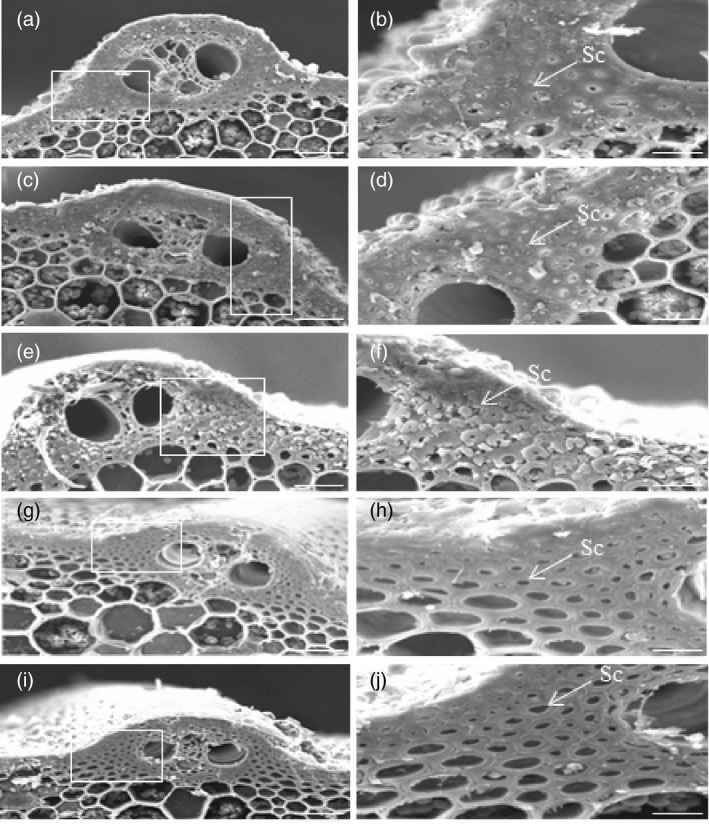
SEM micrographs of the transverse sections of culms showing the state of epidermis and the sclerenchyma cells around the vascular bundle. ZH11 (a, b), OE166b (c, d), RNAiHox32 (e, f), STTM166b (g, h) and OEHox32 (i, j). The white rectangles indicate the parts being magnified in the right panels. Bars = 25 μm (a, c, e, g, i) and 10 μm (b, d, f, g, j). Sc, Sclerenchyma cells.
Figure 3.

TEM micrographs of the transverse sections of flag leaves showing cell walls of the sclerenchyma cells. ZH11 (a, b), OE166b (c, d), RNAiHox32 (e, f), STTM166b (g, h) and OEHox32 (i, j). The white rectangles indicate the parts being magnified in the right panels. Bars = 10 μm (a, c, e, g, i) and 2 μm (b, d, f, h, j). (k) Statistical analysis of the thickness of cell walls. Data are shown as mean ± SE (n = 10), Student’s t‐test (two tailed): *, P < 0.05; **, P < 0.01.
OsmiR166b‐OsHox32 pair manipulates cellulose and lignin contents
The thickness of cell wall is significantly altered in OsmiR166b and OsHox32 transgenic plants. Cellulose, lignin and hemicellulose are three main components of the cell walls, and therefore, we analysed the contents of these compounds in transgenic plants. Contents of hemicellulose in the leaves were not significantly different between the transgenic and wild‐type plants (Figure S4). We then checked the contents of cellulose and lignin in transgenic plants. Cellulose was significantly reduced in the leaves of STTM166b and OEHox32 plants but significantly increased in OE166b and RNAiHox32 plants when compared with that in ZH11 (Figure 4a). Similar results were observed for lignin in the leaves of OsmiR166b and OsHox32 transgenic plants (Figure 4b). To further investigate the deposition of cellulose and lignin in cell walls, we stained transverse sections of leaves from transgenic and wild‐type plants with Calcofluor White Stain and conventional Wiesner histochemical reagent, respectively. Calcofluor White Stain reagent stains cellulose, and the fluorescent signal reflects cellulose content; on the other hand, conventional Wiesner (phloroglucinol‐HCl) reagent is known to react with cinnamaldehyde residues in lignin, and the colour intensity of staining is an approximate indicator of lignin content. As shown in Figure 5, OE166b and RNAiHox32 exhibited stronger fluorescence whereas STTM166b and OEHox32 exhibited weaker fluorescence when compared with the fluorescence observed in ZH11 (Figure 5a–e), indicating different amounts of the cellulose deposited in the cell walls of transgenic plants. Ectopic staining of lignin was also observed in the leaf transverse sections (Figure 5f–j), and the different colour intensities shown in the vascular bundle cells indicated differential lignin deposition in the secondary walls that more in OE166b and RNAiHox32 plants whereas less in STTM166b and OEHox32 plants when compared with that in ZH11.
Figure 4.

Cellulose and lignin contents in the flag leaves of wild‐type and transgenic plants at tillering stage. Cellulose (a) and lignin (b) contents of flag leaves. Data from five biological repeats are shown as mean ± SE. Student’s t‐test. *, P < 0.05; **, P < 0.01.
Figure 5.
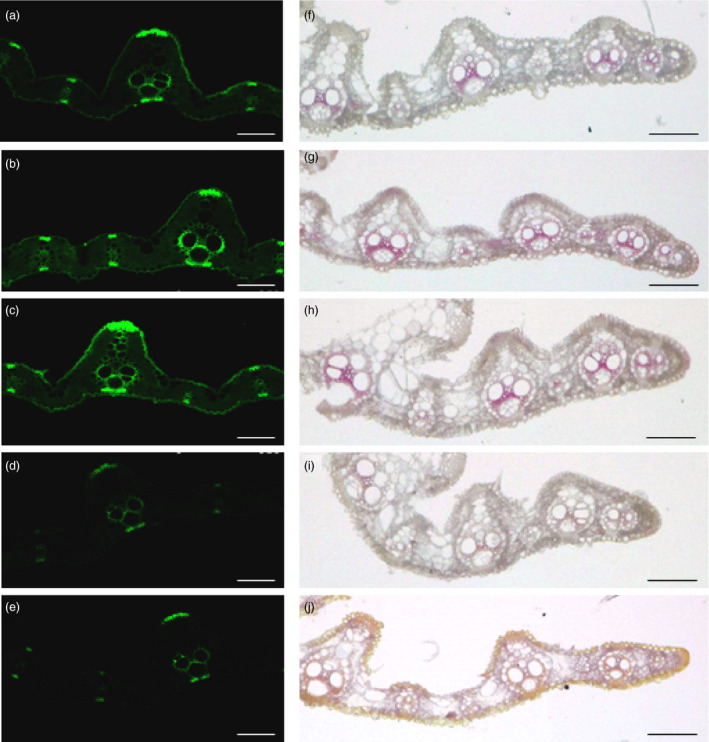
The flag leaf sections stained with Calcofluor White Stain and Wiesner histochemical reagent, respectively, showing cellulose and lignin deposition. (a‐e) Calcofluor White staining exhibits various intensities of green colour proportionally reflecting cellulose deposition in the cells around vascular bundles of ZH11 (a), OE166b (b), RNAiHox32 (c), STTM166b (d) and OEHox32 (e). (f–j) Wiesner histochemical reagent staining shows different coloration in the cells around vascular bundles of ZH11 (f), OE166b (g), RNAiHox32 (h), STTM166b (i) and OEHox32 (j), indicating different lignin deposition. Bars: 100 µm.
Furthermore, we also examined the contents of cellulose and lignin in the culms of transgenic plants. As shown in Figure 6, the contents of cellulose and lignin were significantly increased in OE166b and RNAiHox32 plants but reduced in STTM166b and OEHox32 transgenic plants when compared to those of the wild‐type ZH11. In addition, staining of the transverse sections of culms showed that OE166b and RNAiHox32 transgenic plants accumulated more cellulose and lignin, when compared to the wild‐type ZH11, whereas STTM166b and OEHox32 plants had less cellulose and lignin (Figure 7). These results indicate that the OsmiR166b‐OsHox32 pair regulates the abundance of cellulose and lignin.
Figure 6.

Cellulose and lignin contents in the culms of wild‐type and transgenic plants at heading stage. Cellulose (a) and lignin (b) contents of the second culms counting from the top of rice plants. Data from five biological repeats are shown as mean ± SE. Student’s t‐test. *, P < 0.05; **, P < 0.01.
Figure 7.

The sections of culms stained by Calcofluor White Stain and Wiesner histochemical reagent, respectively, showing cellulose and lignin deposition. (a–e) Calcofluor White staining of the transverse sections of the second culms shows different intensities of green colour in epidermis and vascular bundles of ZH11 (a), OE166b (b), RNAiHox32 (c), STTM166b (d) and OEHox32 (e), indicating different cellulose deposition in the cell walls. (f–j) Wiesner’s staining of the transverse sections of the second culms exhibits various coloration in epidermis and vascular bundles of ZH11 (f), OE166b (g), RNAiHox32 (h), STTM166b (i) and OEHox32 (j), showing different lignin deposition in the cell walls. Bars: 100 µm.
Biosynthesis of cellulose and lignin was controlled by different sets of genes. Expression levels of several genes involved in cellulose and lignin biosynthesis were examined in the wild‐type and transgenic plants (Figure 8). Quantitative RT‐PCR analyses showed that the transcript levels of the cellulose biosynthetic genes OsCESA6 and OsCESA7 and the MLG biosynthetic gene OsCSLF6 (cellulose synthase‐like F6, a major isoform of the ß‐1,3:1,4‐glucan synthase gene) were significantly increased in OE166b and RNAiHox32 plants but decreased in STTM166b and OEHox32 plants when compared with their expression levels in wild‐type ZH11 (Figure 8a). When checked the expression of lignin biosynthesis genes, we found that the expression levels of Os4CL3 and OsCAD2 were significantly increased in OE166b and RNAiHox32 plants but decreased in STTM166b and OEHox32 plants when compared with their expression levels in the wild‐type ZH11. On the other hand, the expression levels of OsPAL1, OsC4H and OsCOMT did not show significant differences between the transgenic and wild‐type plants (Figure 8b). CAD genes function in the last step of monolignol biosynthesis, catalysing hydroxyl‐cinnamyl aldehydes into their corresponding alcohols, and OsCAD2 is regarded as the bona fide CAD gene playing the major role in monolignol biosynthesis (Hirano et al., 2012). The expression of OsCAD2 was further examined in more transgenic lines, and the results showed that the expression of OsCAD2 was significantly regulated in OsmiR166b and OsHox32 transgenic plants (Figure 8c). As a transcription factor, OsHox32 may directly regulate the expression of OsCAD2. Yeast one‐hybrid (Y1H) assay showed that OsHox32 binds to the promoter region of OsCAD2 (Figure 9a). To further confirm the result of Y1H assay, we performed ChIP‐PCR analysis. The results showed the P4 and P5 parts of the promoter were enriched by Flag antibody (Figure 9b, c), indicating that OsHox32 binds to these parts of the OsCAD2 promoter.
Figure 8.
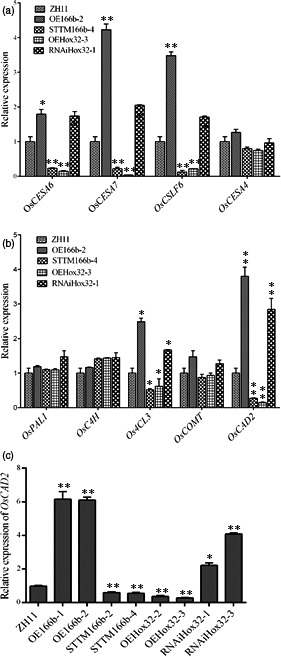
Expression of the cell wall‐related genes. (a) Expression of cellulose and MLG biosynthetic genes in wild‐type and transgenic plants. (b) Expression of lignin biosynthetic genes. (c) Expression of OsCAD2 in different transgenic plants. *, P < 0.05; **, P < 0.01.
Figure 9.

OsHox32 suppresses the expression of OsCAD2 and OsCESA7. (a, d) Y1H assays show OsHox32 binds to the promoters of OsCAD2 (a) and OsCESA7 (d). (b, e) Schematic graphs of the promoter and partial coding region of OsCAD2 (b) and the promoter region of OsCESA7 (e), and the dashed boxes indicate the binding parts tested in ChIP‐PCR. (c, f) ChIP‐PCR confirmed OsHox32 binds to the P4 and P5 parts of OsCAD2 promoter (c) and to the P3, P4 and P9 parts of OsCESA7 promoter (f). The genomic fragment of Actin 1 gene was used as a negative control. Three biological repeats were performed. Student’ t‐test. **, P < 0.01.
OsCESA7 is an essential gene for biosynthesis of the cellulose in the secondary cell walls. Yeast one‐hybrid assay showed that OsHox32 binds to the promoter region of OsCESA7 (Figure 9d), and ChIP‐PCR assay confirmed that OsHox32 binds to the P3, P4 and P9 parts of the OsCESA7 promoter (Figure 9e, f).
OsHox32 interacts with OSH15 and co‐suppresses the expression of OsCAD2
OSH15, a Class I KNOX protein, was reported to bind to OsCAD2 promoter and suppress the expression of OsCAD2, leading to reduced lignin content (Yoon et al., 2017). Yeast two‐hybrid (Y2H) assay showed that OsHox32 interacts with OSH15 (Figure 10a), and bimolecular fluorescence complementation (BiFC) experiment further confirmed the interaction between OsHox32 and OSH15 (Figure 10b). OsHox32 and OSH15 each contain several domains (Figure S5a), and we made the truncated proteins containing different domains for each protein. The Y2H assay showed that the START domain‐containing fragment of OsHox32 interacts with the KNOX box‐containing fragment of OSH15 (Figure S5b). On the other hand, OsHox32 binds to P4 and P5 parts of the OsCAD2 promoter, while OSH15 binds to P5 and P6 parts of the OsCAD2 promoter (Yoon et al., 2017), and the overlap of the binding sites implies the effects of interaction on the gene expression. To investigate the regulatory effects of OsHox32 and OSH15 on the expression of OsCAD2 and OsCESA7, we carried out the dual‐luciferase assay which showed that coexistence of OsHox32 and OSH15 synergistically suppressed the expression of OsCAD2 (Figure 10c, d), but the coexistence did not affect the suppression of OsHox32 on the expression of OsCESA7 (Figure 10c, e).
Figure 10.

OsHox32 interacts with OSH15. (a) Y2H assay shows that OsHox32 interacts with OSH15. T/L, synthetic complete medium lacking Trp and Leu. T/L/H/A, synthetic complete medium lacking Trp, Leu, His and Ade. Triangle indicates 10‐fold dilution of yeasts. Positive control, pGADT7‐7/pGBKT7‐53 combination from Clontech Kit. (b) BiFC confirms the interaction of OsHox32 and OSH15. Bar = 20 μm. (c) Diagrams of the effector and the reporter constructs used in dual‐luciferase assay. (d, e) Dual‐luciferase assay shows that OsHox32 and OSH15 synergistically suppress the expression of OsCAD2 (d), but the coexistence of these two proteins does not have the synergistic effect on the suppression of OsCESA7 (e). Three biological repeats were performed. Student’s t‐test. **, P < 0.01, ns, not significant.
Discussion
MicroRNAs play vital roles in many aspects of plant growth and development by mediating target gene expression. A single miRNA was predicted to target multiple genes, and the best way to elucidate the function of a miRNA is to find out its authentic targets and study the function of the pair of miRNA and target gene. miR166 is conserved in dicots and monocots and forms a big family of 24 members in rice (Oryza sativa L.) (http://structuralbiology.cau.edu.cn/PNRD/). OsmiR166b was reported to be located in a yield‐related QTL interval and differentially expressed in different tissues of an elite hybrid rice (Fang et al., 2013), and it was confirmed to target OsHox32, a HD‐ZIP III family gene. Phenotypic observation of the OsmiR166b and OsHox32 transgenic rice plants demonstrated that the OsmiR166b‐OsHox32 pair regulates plant growth and development and architecture formation.
Zhang et al (2018) reported that knockdown of rice miR166 causes altered stem xylem development and decreased hydraulic conductivity, which leads to rolled leaf and thus confers drought resistance. Li et al (2016) found that overexpression of OsHox32 results in narrow leaves and reduced plant height. In this study, we observed rolled leaves and reduced plant height in STTM166b and OEHox32 transgenic plants, which is in agreement with their findings. However, we also observed that STTM166b and OEHox32 transgenic plants displayed drooping leaf and brittle culms, which implies that the OsmiR166b‐OsHox32 module plays important roles not only in controlling growth and development but also in modulating the mechanical strength of plants.
Mechanical strength of plants is determined by the components of cell walls. SEM and TEM analyses demonstrated that the thickness of cell walls of OsmiR166b and OsHox32 transgenic plants was different to that of WT. Mixed‐linkage glucan (MLG) is a polysaccharide and deposited in the primary cell wall to regulate cell wall expansion of the young and expanding tissues, and CSLF6 has a predominant and nonredundant role in the biosynthesis and accumulation of MLG (Kim et al., 2018; Vega‐Sánchez et al., 2012). The cslf6 knockout rice mutant not only showed a decrease in both plant height and stem diameter but also exhibited a significant reduction in culm strength (Vega‐Sánchez et al., 2012). Cellulose biosynthesis is controlled by a set of CESA genes, and mutations in these genes result in changes in cellulose content and defects in plant growth and development. OsCESA6, partially redundant to OsCESA3, is responsible for cellulose biosynthesis in the primary cell wall (Wang et al., 2010). Previous studies showed that cellulose deficiency in primary cell wall is correlated with cell elongation defects and morphology changes (Paredez et al., 2006, Park and Cosgrove, 2012). The reductions in plant height and mechanical strength were also observed in OsmiR166b and OsHox32 transgenic plants, and these phenotypic changes may be partially due to the different expression of OsCESA6 and OsCSLF6. Calcofluor staining assay showed much more cellulose deposition in OE166b and RNAiHox32 transgenic plants but less in STTM166b and OEHox32 plants. CESA4 and 7 are required for cellulose biosynthesis during the secondary cell wall formation. Although the expression of OsCESA4 was not significantly different between the wild‐type and transgenic plants, the expression level of OsCESA7 was significantly regulated by OsmiR166b‐OsHox32 pair, and molecular analyses further confirmed that OsHox32 binds to the promoter region of OsCESA7 and suppresses its expression. The regulation of OsCESA7 may lead to differential deposition of cellulose in the secondary cell walls of the transgenic plants.
In the generally accepted model for secondary cell wall, lignin is proposed to give the wall rigidity and resistance to compressive force (Turner and Somerville, 1997). Lignin biosynthesis genes and pathways have been uncovered and well defined (Raes et al., 2003). CADs catalyse the last step of lignin biosynthesis, and among the 12 CAD genes present in the rice genome, OsCAD2 is the major one responsible for monolignol biosynthesis in rice culm (Hirano et al., 2012). OsCAD2‐mutant gh2 showed reddish‐brown panicles and internodes and exhibited a decrease in lignin content in cell walls when compared to wild‐type plants (Zhang et al., 2006). Besides the gold hull and internode, the oscad2 rice mutant showed brown midrib and decreased lignin content as well (Koshiba et al., 2013). In addition, the maize (Zea mays L.) bm1 (brown midrib 1) mutant harbours mutations in the CAD gene which leads to a decrease in CAD activity and a reduction in lignin content (Halpin et al., 1998). On the other hand, the maize bm3 mutants showed structural changes in COMT gene and exhibited only 10% COMT activity of the WT (Vignols et al., 1995). For lignin biosynthesis, PAL1, C4H and 4CL3 are responsible for the first three steps of the biosynthesis pathway, whereas CAD and COMT account for the rear steps of the pathway (Li et al., 2015; Zhao, 2016). The reduced CAD or COMT activity found in either rice gh2 mutant or maize bm mutants may feedback to the upstream steps of the biosynthesis pathway to stimulate the expression of genes such as 4CL3, leading to the flux of intermediate products to the flavonoid biosynthesis branch that increases flavonoid contents and results in pigment formation in tissues. In this study, OsmiR166b‐OsHox32 pair significantly down‐regulated the expression of OsCAD2 and Os4CL3 in STTM166b and OEHox32 plants, which slows down the biosynthesis speed of the whole lignin pathway and does not mediate the flux of intermediate products to flavonoid biosynthesis, therefore leading to less lignin accumulation and no pigment formation in the midribs and internodes of STTM166b and OEHox32 plants.
The OsmiR166b‐OsHox32 pair regulates lignin biosynthesis through the binding of OsHox32 to the promoter of OsCAD2. Generally, the lignin biosynthetic genes are regulated by a pyramid‐shaped regulatory hierarchy of transcription factors (Nakano et al., 2015; Zhao and Dixon, 2011). NAC and MYB transcription factors have been demonstrated to act as master switches of the entire secondary cell wall biosynthesis. SND1 is the first‐layer master switch for secondary cell wall biosynthesis and controls the second‐layer master switches of MYB46 and MYB83 by binding to their promoters (McCarthy et al., 2009). Except for this general transcriptional cascade, other ways are also found to be involved in the secondary cell wall biosynthesis. For example, lignin laccase genes were demonstrated to be the targets of miR397, and the miR397‐mediated cleavage of laccase transcripts leads to reduced lignin content (Lu et al., 2013; Wang et al., 2014). OSH15 is a member of the knotted1‐type homeobox gene family, and mutations in this gene caused a dwarf phenotype and an increased deposition of lignin in internodes (Sato et al., 1999; Yoon et al., 2017). Molecular investigation showed that OSH15 is a negative regulator of lignin biosynthesis as it binds to the promoter region of OsCAD2 and suppresses the expression (Yoon et al., 2017). OsHox32 acts as another negative regulator of OsCAD2 by binding to the promoter and suppressing the expression of OsCAD2. In this case, OsCAD2 is regulated by two negative factors, and the binding sites for these two proteins, OsHox32 and OSH15, are overlapped on the promoter region. OsHox32 interacts with OSH15, and the interaction enhances the suppression of OsCAD2. Since OsCAD2 is the bona fide CAD gene that plays important roles in monolignol biosynthesis (Hirano et al., 2012), regulation of OsCAD2 expression may act as the key switch to control lignin content. Overexpression of OsHox32 not only increases suppression of OsCAD2 by OsHox32 itself but also enhances the suppression of OsCAD2 by OSH15, thus greatly reduces lignin content in tissues. However, lignin is essential for plant growth and development, and sometimes suppression of OsCAD2 should be released or controlled to some extent. In this respect, OsmiR166b exerts power to affect lignin biosynthesis by cleaving OsHox32 transcript and releasing the OsHox32 suppression on OsCAD2. That the OsmiR166b‐OsHox32 pair regulates lignin biosynthesis represents an elaborate way that is more efficient than the direct way exemplified by miR397 and OSH15‐mediated regulation. The diversity of the regulatory pathways might be dominant in a context‐dependent manner, but how these pathways corporately regulate lignin biosynthesis needs further investigation.
Methods
Plant materials and growth conditions
All transgenic plants were generated from rice (Oryza sativa L.) Zhonghua 11 (japonica cv. ZH11). Multiple transgenic plants were obtained, and two representative homozygous lines for each gene transformation were selected to use in this study. Rice plants including transgenic lines and wild‐type ZH11 plants were grown in a paddy field under natural conditions during summer and autumn seasons at South China Botanical Garden, Guangzhou, China, and treated with normal management.
Plasmid constructs and rice transformation
To specifically block the function of OsmiR166b, we applied the short tandem target mimic (STTM) method (Yan et al., 2012), in which the STTM module contained two copies of OsmiR166b that were linked by a 48 nt spacer. Overexpression of miR166b (OE166b) was constructed by inserting the PCR‐amplified miR166b precursor fragment into pCAMBIA1301 vector and expressed under control of the 35S promoter.
The full‐length cDNA fragment of OsHox32 was amplified and inserted into pCAMBIA1301 vector and expressed under control of the 35S promoter. The resulting construct was used to overexpress OsHox32 in rice plants. To generate OsHox32‐RNAi transgenic plants, we PCR‐amplified a 214 bp fragment specific to the first exon of the OsHox32 gene and inserted it into pTCK303 vector in opposite directions. The resulting construct was used to suppress the expression of OsHox32 in rice plants.
All constructs were individually transformed into Agrobacterium tumefacines strain EHA105, and the transformed EHA105 strains were used to transform rice calli to generate transgenic plants. Totally, we got 11, 18, 17 and 15 transgenic lines for STTM166b, OE166b, RNAiHox32 and OEHox32, respectively. The primer sequences used for plasmid construction are listed in supporting information Table S2.
Gene expression analysis
Total RNAs were extracted from leaves collected from plants at tillering stage, and the first‐strand cDNAs were generated from equal amounts of total RNAs using a reverse transcription kit (TaKaRa, Shiga, Japan). Quantitative reverse transcription PCR was performed with three biological repeats on a Roche Light Cycler 480 real‐time PCR machine with SYBR Premix ExTaq II (TaKaRa). PCRs were carried at 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s, and 60 °C for 10 s, and then 72 °C for 15 s. The rice Actin1 gene was used as the internal control, and relative expression levels of genes were calculated using the 2−∆∆CT method (Livak and Schmittgen, 2001). Primer sequences are listed in Table S2.
Protein–protein interaction analysis
For yeast two‐hybrid (Y2H) analysis, the full coding sequence fragment of OsHox32 or OSH15 was accordingly cloned into the pGBKT7 or pGADT7 vector. In addition, the truncated OsHox32 fragments containing either the HD domain or the START domain were accordingly cloned into the above vectors as well. On the other hand, the truncated OSH15 fragments containing either the KNOX domain or the ELK or the HD domain were cloned into one of the above vectors. Various combinations of different constructs were tested for protein–protein interaction in yeasts. The Y2H assay was performed following the manufacturer’s instructions (Clontech, Palo Alto, CA).
For bimolecular fluorescence complementation (BiFC) analysis, the full‐length fragment of the OsHox32 (without stop codon) coding region was cloned into pUC‐SPYNE to generate OsHox32‐nYFP fusion protein. On the other hand, the full‐length fragment of the OSH15 coding region (without stop codon) was cloned into pUC‐SPYCE vector to generate OSH15‐cYFP fusion protein. These two constructs were combined to transfect Arabidopsis protoplast cells, and the cells were then incubated at 23 °C for 16 h. Yellow fluorescent protein (YFP) fluorescence was checked by DMI6000B inverted fluorescence microscope (Leica, Wetzlar, Germany) under DAF channel. Primer sequences are listed in Table S2.
ChIP‐PCR assay
Plasmid DNAs of OsHox32‐3flag and pCactF‐3flag were used to transfect rice protoplast cells (Zhang et al., 2011). The transfected cells were incubated at 26°C for 12 h and then cross‐linked with 1% formaldehyde solution (in 1× PBS buffer with pH 7.4). Chromatins were isolated and sheared to an average length of 500 bp by sonication. The solubilized chromatins were immunoprecipitated with anti‐flag antibody. The cross‐linking was then reversed, and the co‐immunoprecipitated DNAs were then recovered and analysed by quantitative PCR with SYBR Premix ExTaq II (TaKaRa) using gene‐specific primers. ChIP assays were performed as described previously (Lee et al., 2017). Sequences of the primers are listed in Table S2
Yeast one‐hybrid assay
For yeast one‐hybrid (Y1H) analysis, the 146 bp fragment of the OsCAD2 promoter region (from −593 to −447 upstream of the start codon) and 119 bp fragment of the OsCESA7 promoter region (from −1674 to −1555 upstream of the start codon) were PCR‐amplified and inserted into the pAbAi vector, respectively. The full‐length cDNA of OsHox32 was cloned into pGADT7 vector. Yeast transformation and assay were performed following the manufacturer’s instructions (Clontech, Mountain View, CA). Primer sequences are listed in Table S2.
Dual‐luciferase assay
For dual‐luciferase reporter assays, the coding sequences of OsHox32 and OSH15 were individually PCR‐amplified and cloned into the pGreenII 62‐SK vector as effectors, whereas the promoter region of OsCAD2 (−2500 bp upstream of ATG), were inserted into pGreenII 0800‐LUC vector as a reporter. The reporter and effectors were co‐transfected into Arabidopsis protoplast cells in different combinations and incubated at 23 °C for 12 h. Firefly LUC and REN activities were measured by a multimode reader (infinite M200PRO) using the Dual‐Luciferase Reporter Assay System (Promega, Fitchburg, WI, USA). The parameter used for measurement was chemiluminescence, and integration time was 5000 ms. Three biological repeats were performed.
Scanning electron microscopy
The central parts of the second internodes from the top of rice plants at heading stage were collected and excised into small pieces with a razor, and the pieces were immediately placed in the solution of 70% ethanol, 5% acetic acid and 3.7% formaldehyde for 18 h. Samples were critical point dried, sputter‐coated with gold in an E‐100 ion sputter (Mito City, Japan) and observed with a scanning electron microscope (JSM‐6360LV, Japan).
Transmission electron microscopy
The central parts of flag leaf blade at tillering stage were harvested and excised into small pieces, which were then fixed in the solution of 2.5% (w/v) glutaraldehyde in 0.1 m PBS (4 mm sodium phosphate, pH 7.2; 200 mm NaCl) at 4 °C for overnight. Samples were dehydrated through a gradient of ethanol and embedded with Spur Kit (Sigma, Darmstadt, Germany). The 80‐nm ultrathin sections were prepared with an Ultracut E ultramicrotome (Leica) and picked up on formvar‐coated copper grids. After post‐staining with uranyl acetate and lead citrate, the specimens were observed under a transmission electron microscope (JEM‐1010, Japan).
Cellulose and lignin histochemical staining
For cellulose staining, paraffin‐embedded transverse sections (20 μm thickness) of the central parts of the flag leaf blade at tillering stage and of the central parts of the second culms at heading stage were stained with a drop of Calcofluor White Stain (Sigma, Darmstadt, Germany) and a drop of 10% KOH for 1 min and then visualized with a fluorescent microscope (Leica). Images were captured GFP excitation, a range of 375–525 nm.
For histochemical localization of lignin, materials from the same parts of leaves and culms at the same stage as those used for cellulose staining were used. Paraffin‐embedded transverse sections (20 μm thickness) of leaves and culms were stain with phloroglucinol‐HCl solution for 2 min in phloroglucinol solution (5% in ethanol: water [95:5, v/v]; 36% HCl) to distinguish the lignified cell walls (Anderson et al., 2015), and then, sections were photographed using LEICA DVM6 stereomicroscope.
Cellulose and lignin measurement
To measure cellulose and lignin contents, flag leaves of rice plants at tillering stage and the second culms from the top of rice plants at heading stage were harvested, respectively, and then prepared into alcohol‐insoluble residue (AIR) (Harholt et al., 2006). Tissues were ground into fine power in liquid nitrogen, dissolved with 80% ethyl alcohol and then boiled at 100 °C for 20 min. After cooling to room temperature, samples were centrifuged at 2000 g for 10 min and the supernatant was removed. The residues were resuspended in 80% ethyl alcohol, then in 80% acetone, and then in 100% acetone, and finally air‐dried. The air‐dried residues were treated with pullulanase M3 (0.5 U/mg, Megazyme) and alpha‐amylase (0.75 U/mg, Sigma) in 0.1 m NaOAc buffer (pH 5.0) for overnight, and then centrifuged at 2000 g for 10 min, the residues were washed with 80% ethyl alcohol and then dried at 35°C for overnight to get AIR.
The cellulose content was measured using a modified method as described by Updegraff (1969). In brief, 100 mg AIR were hydrolysed with 3 mL Updegraff reagent (Acetic acid: HNO3 : ddH2O = 8:1:2) by boiling at 100°C in a water bath for 30 min and then cooled to room temperature. After centrifugation at 2000 g for 10 min, the supernatant was removed, and the residues were washed with distilled water once and then hydrolysed with 67% sulphuric acid for 1 h. Samples were then centrifuged at 2000 g for 10 min. Use 1 mL supernatant and dilute to 100 mL with distilled water. Then adding 10 mL ice‐cold anthrone reagent to solution and then boiling at 100 °C in a water bath for 15 min. After that, samples were cooled on ice for 2 min and then left at room temperature for 10 min. Finally, samples were read at 620 nm using a spectrophotometer (UV‐1800).
The lignin content was quantified according to the method described by Fukushima and Hatfield (2001). Briefly, 100 mg AIR were hydrolysed with 4 ml 25% acetyl bromide at 50 °C in a water bath for 2 h, then adding acetic acid to bring the volume to 16 mL. Then mixed 0.5 mL reaction solution with 7.5 mL acetic acid, 1.5 mL 0.3 m NaOH and 0.5 mL 0.5 m hydroxylamine hydrochloride at room temperature for 30 min. Samples were read at 280 nm using a spectrophotometer (UV‐1800).
Conflict of interests
The authors declare no competing interests.
Authors’ contributions
H.C. performed most experiments, including vector constructions, PCR, protein–protein interaction, ChIP‐PCR, dual‐luciferase assay, histochemical staining, cellulose and lignin content measurement, R.Q.F. completed STTM construction and obtained STTM166b transgenic plants. R.F.D. helped in SEM and TEM. J.X.L. conceived and supervised the project. H.C. and J.X.L. analysed data and wrote the manuscript.
Supporting information
Figure S1 OsmiR166b targets OsHox32
Figure S2 Characterization of transgenic plants
Figure S3 Comparison of the internodes and leaves from ZH11 and transgenic plants
Figure S4 Hemicellulose content of the flag leaves from wild‐type and transgenic plants at tillering stage
Figure S5 Interaction between the truncated proteins of OsHox32 and OSH15
Table S1 Agronomic trait investigation for the OsmiR166b and OsHox32 transgenic rice plants.
Table S2 Sequences of primers used in this study.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31970523, 31671271).
Chen, H. , Fang, R. , Deng, R. and Li, J. (2021) The OsmiRNA166b‐OsHox32 pair regulates mechanical strength of rice plants by modulating cell wall biosynthesis. Plant Biotechnol. J., 10.1111/pbi.13565
References
- Anderson, N.A. , Tobimatsu, Y. , Ciesielski, P.N. , Ximenes, E. , Ralph, J. , Donohoe, B.S. , Ladisch, M. and et al. (2015) Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell, 27, 2195–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava, A. , Mansfield, S.D. , Hall, H.C. , Douglas, C.J. and Ellis, B.E. (2010) MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis in florescence stem. Plant Physiol. 154, 1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. and Jarvis, M.C. (2012) Comparative structure and biomechanics of plant primary and secondary cell walls. Front. Plant Sci. 3, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhiti, M. and Stasolla, C. (2009) Structure and function of homodomain‐leucine zipper (HD‐Zip) proteins. Plant Signal Behav. 4, 86–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, H. , Yamaguchi, M. , Tamura, T. , Nakano, Y. , Nishikubo, N. , Yoneda, A. , Kato, K. et al. (2015) Multiple classes of transcription factors regulate the expression of VASCULAR‐RELATED NAC‐DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. [DOI] [PubMed] [Google Scholar]
- Fang, R. , Li, L. and Li, J.X. (2013) Spatial and temporal expression modes of microRNAs in an elite rice hybrid and its parental lines. Planta, 238, 259–269. [DOI] [PubMed] [Google Scholar]
- Fukushima, R.S. and Hatfield, R.D. (2001) Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. J. Agric. Food Chem. 7, 3133–3139. [DOI] [PubMed] [Google Scholar]
- Gao, J. , Chen, H. , Yang, H.F. , He, Y. , Tian, Z.H. and Li, J.X. (2018) A brassinosteroid responsive miRNA‐target module regulates gibberellin biosynthesis and plant development. New Phytol. 220, 488–501. [DOI] [PubMed] [Google Scholar]
- Halpin, C. , Holt, K. , Chojecki, J. , Oliver, D. , Chabbert, B. , Monties, B. , Edwards, K. et al. (1998) Brown‐midrib maize (bm1): a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14, 545–553. [DOI] [PubMed] [Google Scholar]
- Harholt, J. , Jensen, J.K. , Sorensen, S.O. , Orfila, C. , Pauly, M. and Scheller, H.V. (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic Arabinan in Arabidopsis . Plant Physiol. 140, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, K. , Aya, K. , Kondo, M. , Okuno, A. , Morinaka, Y. and Matsuoka, M. (2012) OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep. 31, 91–101. [DOI] [PubMed] [Google Scholar]
- Kim, S.J. , Starla, Z.D. , Jensen, J.K. , Wilkerson, C.J. , Keegstra, K. and Brandizzi, F. (2018) In the grass species Brachypodium distachyon, the production of mixed‐linkage (1,3;1,4) ‐β‐glucan (MLG) occurs in the Golgi apparatus. Plant J. 93, 1062–1075. [DOI] [PubMed] [Google Scholar]
- Koshiba, T. , Murakami, S. , Hattori, T. , Mukai, M. , Takahashi, A. , Miyao, A. , Hirohiko Hirochika, H. et al. (2013) CAD2 deficiency causes both brown midrib and gold hull and internode phenotypes in Oryza sativa L. cv. Nipponbare. Plant Biotechnol. 30, 365–373. [Google Scholar]
- Kubo, M. , Udagawa, M. , Nishikubo, N. , Horiguchi, G. , Yamaguchi, M. , Ito, J. , Mimura, T. et al. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. , Jin, S. , Kim, S.Y. , Kim, W. and Ahn, J.H. .(2017) A fast, efficient chromatin immunoprecipitation method for studying protein‐DNA binding in Arabidopsis mesophyll protoplasts. Plant Methods, 13, 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.X. , Yu, S.B. , Xu, C.G. , Tan, Y.F. , Gao, Y.J. , Li, X.H. and Zhang, Q.F. (2000) Analyzing quantitative trait loci for yield using a vegetatively replicated F2 population from a cross between the parents of an elite rice hybrid. Theor. Appl. Genet. 101, 248–254. [Google Scholar]
- Li, X.J. , Yang, Y. , Yao, J.L. , Chen, G.X. , Li, X.H. , Zhang, Q.F. and Wu, C.Y. (2009) FLEXIBLE CULM 1 encoding a cinnamyl‐alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol. Biol. 69, 685–697. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Kim, J.I. , Pysh, L. and Chapple, C. .(2015) Four isoforms of Arabidopsis 4‐Coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169, 2409–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Qian, Q. , Zhou, Y. , Yan, M. , Sun, L. , Zhang, M. , Fu, Z. et al. (2003) BRITTLE CULM1, which encodes a COBRA‐like protein, Affect the mechanical properties of rice plants. Plant Cell, 15, 2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.Y. , Shen, A. , Xiong, W. , Sun, Q.L. , Luo, Q. , Song, T. , Li, Z.L. et al. (2016) Overexpression of OsHox32 results in pleiotropic effects on plant type architecture and leaf development in rice. Rice, 9, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐∆∆CT Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, S.F. , Li, Q.Z. , Wei, L.R. , Chang, M.J. , Tunlaya‐Anukit, S. , Kim, H. , Liu, J. et al. (2013) Ptr‐miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl Acad. Sci. USA, 110, 10848–10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, R.L. , Zhong, R. and Ye, Z.H. (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. [DOI] [PubMed] [Google Scholar]
- Mele, G. , Ori, N. , Sato, Y. and Hake, S. .(2003) The knotted1‐like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17, 2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, Y. , Yamaguchi, M. , Endo, H. , Rejab, N.A. and Ohtani, M. (2015) NAC‐MYB‐based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez, A.R. , Somerville, C.R. and Ehrhardt, D.W. (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science, 312, 1491–1495. [DOI] [PubMed] [Google Scholar]
- Park, Y.B. and Cosgrove, D.J. (2012) Changes in cell wall biomechanical properties in the xyloglucan‐deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol. 158, 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S. , Cassman, K.G. , Virmani, S.S. , Sheehy, J. and Khush, G.S. (1999) Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop Sci. 39, 1552–1559. [Google Scholar]
- Raes, J. , Rohde, A. , Christensen, J.H. , Peer, Y.V. and Boerjan, W. (2003) Genome‐wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W. , Reinhart, B.J. , Lim, L.P. , Burge, C.B. , Bartel, B. and Bartel, D.P. .(2002) Prediction of plant microRNA targets. Cell, 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Sasaki, A. , Ashikari, M. , Ueguchi‐Tanaka, M. , Itoh, H. , Nishimura, A. , Swapan, D. , Ishiyama, K. et al. (2002) A mutant gibberellin‐synthesis gene in rice. Nature, 416, 701–702. [DOI] [PubMed] [Google Scholar]
- Sato, Y. , Sentoku, N. , Miura, Y. , Hirochika, H. , Kitano, H. and Matsuoka, M. (1999) Loss‐of‐function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18, 992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K. , Murata, K. , Yamazaki, M. , Onosato, K. , Miyao, A. and Hirochika, H. (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 133, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor‐Teeples, M. , Lin, L. , de Lucas, M. , Turco, G. , Toal, T.W. , Gaudinier, A. et al. (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature, 517, 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R. and Somerville, C.R. (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell, 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff, D.M. (1969) Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424. [DOI] [PubMed] [Google Scholar]
- Vega‐Sanchez, M.E. , Verhertbruggen, Y. , Christensen, U. , Chen, X.W. , Sharma, V. , Varanasi, P. , Jobling, S.A. et al. (2012) Loss of cellulose synthase‐like f6 function affects mixed‐linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignols, F. , Rlgau, J. , Torres, M.A. , Capellades, M. and Puigdomènech, P. (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O‐methyltransferase. Plant Cell, 7, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.Y. , Zhang, S.C. , Yang, Y. , Luo, Y.C. , Liu, Q. , Ju, C.L. , Zhang, Y.C. et al. (2014) MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 12, 1132–1142. [DOI] [PubMed] [Google Scholar]
- Wang, L.Q. , Guo, K. , Li, Y. , Tu, Y.Y. , Hu, H.Z. , Wang, B.R. , Cui, X.C. et al. (2010) Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J. , Gu, Y. , Jia, X. , Kang, W. , Pan, S. , Tang, X. , Chen, X. et al. (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell, 24, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J. , Cho, L.H. , Antt, H.W. , Koh, H.J. and An, G. (2017) KNOX Protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 174, 312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K. , Sakamoto, S. , Kawai, T. , Kobayashi, Y. , Sato, K. , Ichinose, Y. , Yaoi, K. et al. (2013) Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front. Plant Sci. 4, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.H. , Zhang, A.K. , Srivastava, P.Y. , Bai, J. , Fang, J. , Shi, H. and Zhu, J.K. (2018) Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 176, 2082–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Qian, Q. , Huang, Z. , Wang, Y. , Li, M. , Hong, L. , Zeng, D. et al. (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl‐alcohol dehydrogenase in rice. Plant Physiol. 140, 972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Su, J.B. , Duan, S. , Ao, Y. and Wang, H.B. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast‐related processes. Plant Methods, 7, 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q. (2016) Lignification: flexibility, biosynthesis and regulation. Trends Plant Sci. 21, 13–721. [DOI] [PubMed] [Google Scholar]
- Zhao, Q. and Dixon, R.A. (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16, 227–233. [DOI] [PubMed] [Google Scholar]
- Zhong, R. , Lee, C. , McCarthy, R.L. , Reeves, C.K. , Jones, E.G. and Ye, Z.H. (2011) Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 OsmiR166b targets OsHox32
Figure S2 Characterization of transgenic plants
Figure S3 Comparison of the internodes and leaves from ZH11 and transgenic plants
Figure S4 Hemicellulose content of the flag leaves from wild‐type and transgenic plants at tillering stage
Figure S5 Interaction between the truncated proteins of OsHox32 and OSH15
Table S1 Agronomic trait investigation for the OsmiR166b and OsHox32 transgenic rice plants.
Table S2 Sequences of primers used in this study.


