Fig. 1.
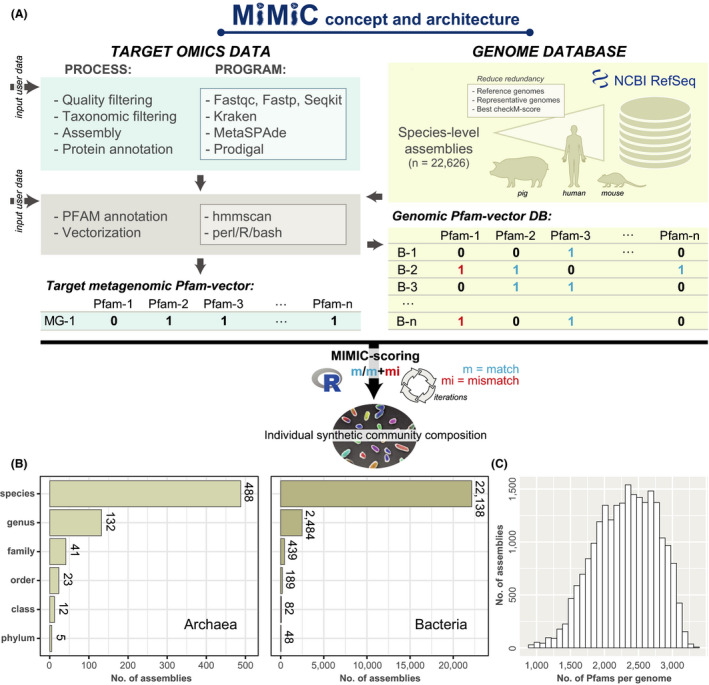
Schematic overview of the architecture and content of MiMiC. See methods section for all details.
A. (Meta)genomic reads are quality‐checked and processed into binary vectors of protein families (Pfams). The resulting metagenomic profile (MG‐1) is used for iterative selection of a minimal number of genomes (calculated by a knee point approach) from the database (B‐1 to B‐n) that best cover the functional potential of the input data (highest number of matches; least number of mismatches).
B. The genome database currently consists of 22 627 species‐level genomes of archaea and bacteria spanning a total of 53 phyla, with an average of approximately 2500 Pfams per genome.
