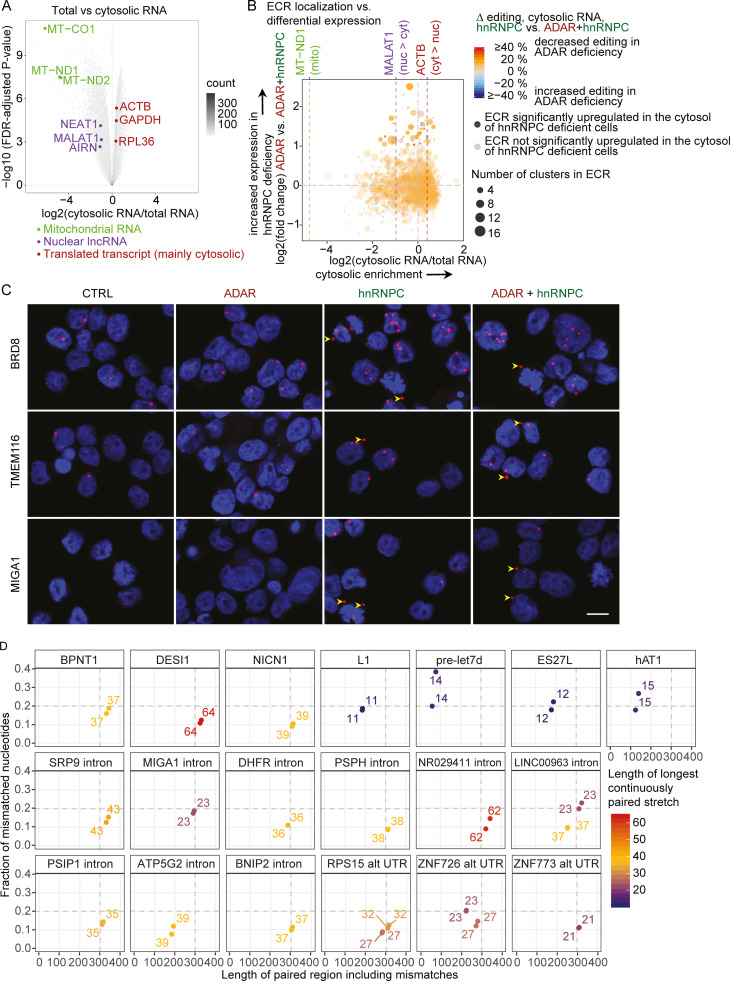
Figure 9.
hnRNPC deficiency–dependent up-regulated editing cluster groups gain access to the cytosol.(A) Differential expression analysis of genes between total cellular RNA and RNA from cytosolic extracts (see Materials and methods) of THP-1 (WT clone) as indicated in Materials and methods. Mitochondrially encoded genes (green), nuclear lncRNAs (purple), and expressed housekeeping mRNAs are highlighted; all else are displayed as 2D-density plot. (B) Four-way plot of log2 (fold-change) expression of A-to-I ECRs. y axis: expression in ADAR gRNA singly versus ADAR+hnRNPC gRNA doubly nucleofected cells on day 4 after nucleofection. x axis: log2 (fold change) between total RNA and RNA from cytosolic extracts. Purple lines indicate values for MT-ND1, MALAT1, and ACTB RNAs as in A. Circle color indicates the average reduction in editing in hnRNPC single versus hnRNPC+ADAR double-deficiency as difference of the average percent editing across all editing sites within each cluster group. Average of two samples per condition. Transparency indicates significant up-regulation in hnRNPC+ADAR over ADAR gRNA nucleofected cells; size of circles indicates number of clusters per cluster group. (A and B) Data are from three independent experiments. (C) BaseScope analysis (red) of select ECRs in THP-1 with the indicated deficiencies. Arrowheads point to cytosolic transcripts. Nuclei were stained with DAPI (blue). Images are details of images in Fig. S4 B, which are representatives of three images each from two independent experiments. Scale bar, 10 µm. (D) Comparison of previously described MDA5 ligands (BPNT1, DESI1, and NICN1) and negative controls (hAT1, L1, pre-let7d, and ES27L) to putative MDA5 ligands within ECRs by indicating length, including mismatches of each strand of the dsRNA stretch, fraction of mismatched nucleotides, and length of the longest uninterrupted dsRNA stretch within putative MDA5 ligands. CTRL, nontarget control; cyt, cytosolic; 2D, two-dimensional; lncRNA, long noncoding RNA; mito, mitochondrially encoded; nuc, nuclear.