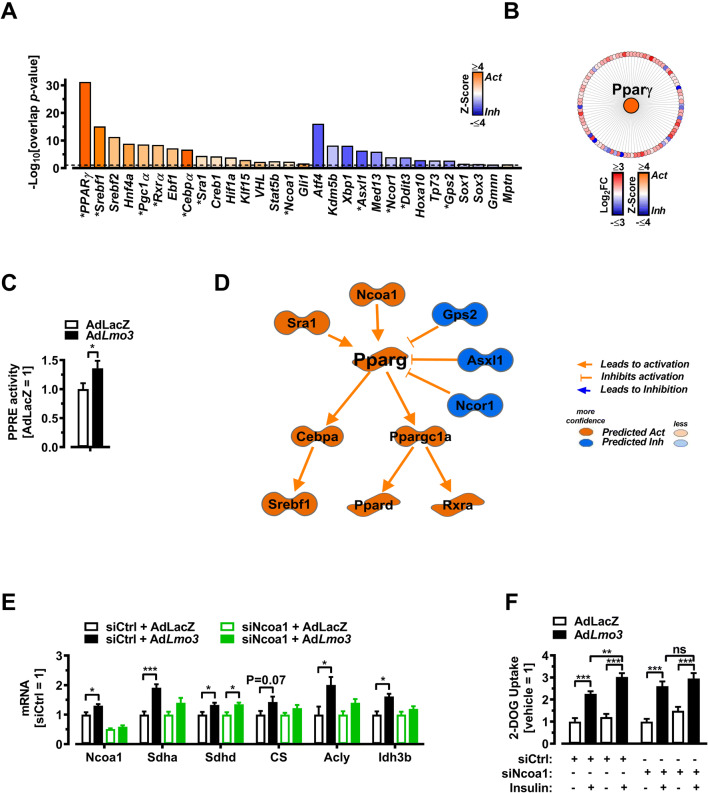
Fig. 6.
LMO3 increases mitochondrial oxidative gene expression and glucose uptake via NCoA1/SRC-1. A IPA-predicted upstream regulators from AdLmo3- versus AdLacZ-transduced 3T3-L1 adipocytes showing activation Z-score (bars). Top-15 transcriptional regulators with an overlap p < 0.05 by IPA (see “Methods” for description) were predicted to be upstream regulators. *indicates genes comprising the IPA-predicted mechanistic network displayed in (D). B IPA-predicted upstream regulators (center, colored by Z-score) and target genes (outer circle, colored by Fold Change) in AdLmo3- versus AdLacZ-transduced 3T3-L1 adipocytes. C ELISA-based PPARγ transcription reporter assay demonstrating that LMO3 activated PPARγ transcriptional activity in mature adipocytes compared with LacZ controls (n = 6). D IPA-predicted mechanistic network based on upstream regulators as shown in (F). See supplemental information for gene symbol description. See Figure S1B for explanation of molecule shapes. E Q-PCR analysis of selected “TCA cycle” genes in siCtr- or siNcoa1-transfected control (AdLacZ-) or LMO3-overexpressing (AdLmo3) 3T3-L1 adipocytes (n = 5). F Glucose uptake in in siCtr- or siNcoa1-transfected control (AdLacZ-) or LMO3-overexpressing 3T3-L1 adipocytes (n = 5). After 4-h serum starvation, the cells received mock or insulin treatment for 20 min for measurement of 2-DG uptake. *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant