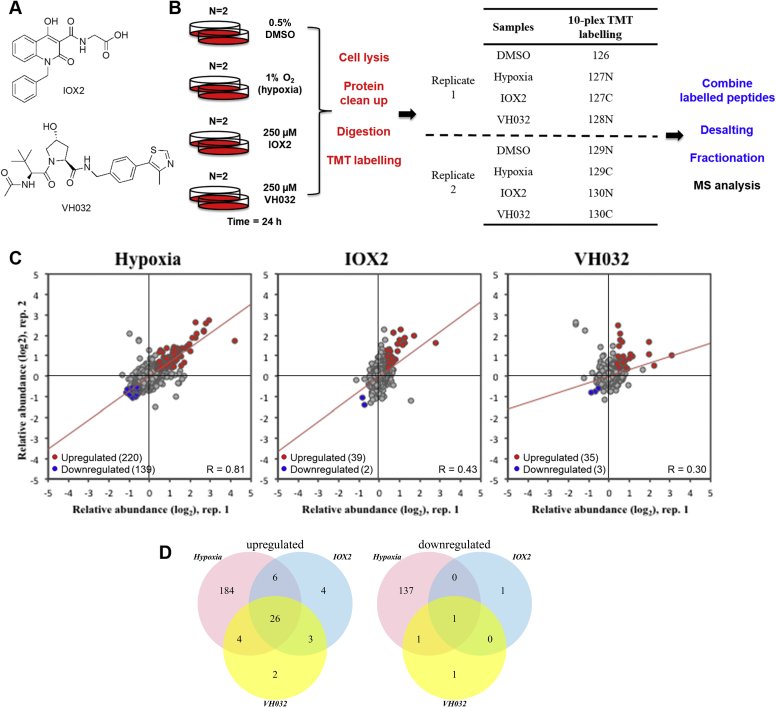
Figure 1.
Unbiased proteomics identify VHL protein level increase upon VHL inhibitor treatment.A, chemical structures of IOX2 and VH032. B, diagram depicting the workflow of tandem mass tag (TMT) labeling. HeLa cells were treated with 0.5% DMSO, 1% O2 (hypoxia), 250 μM IOX2, and 250 μM VH032 for 24 h—in biological duplicate. Proteins were obtained by cell lysis, cleaned up, digested by trypsinization, and labeled with 10-plex TMT labeling reagent. Labeled peptides were combined, desalted, fractionated, and analyzed by MS. Red text indicates steps carried out at the protein level, and blue text indicates steps performed on peptides. C, scatter plot representation of relative protein abundances obtained for different treatment conditions compared with respective replicate of vehicle (DMSO)-treated cells, for a total of 8043 proteins quantified. The two axes are relative abundance (log2FC) from two different replicates in this experiment. Red dots represent upregulated genes in both replicates (absolute fold-change difference to DMSO > 1.3), and blue dots represent downregulated genes in both replicates (absolute fold-change difference to DMSO < 0.7). Red line is the linear fit to the data. D, Venn diagrams depicting the number of upregulated genes and downregulated genes in the two replicates comparing the presence of hypoxia, IOX2, or VH032 to DMSO control. DMSO, dimethyl sulfoxide; VHL, Von Hippel–Lindau.