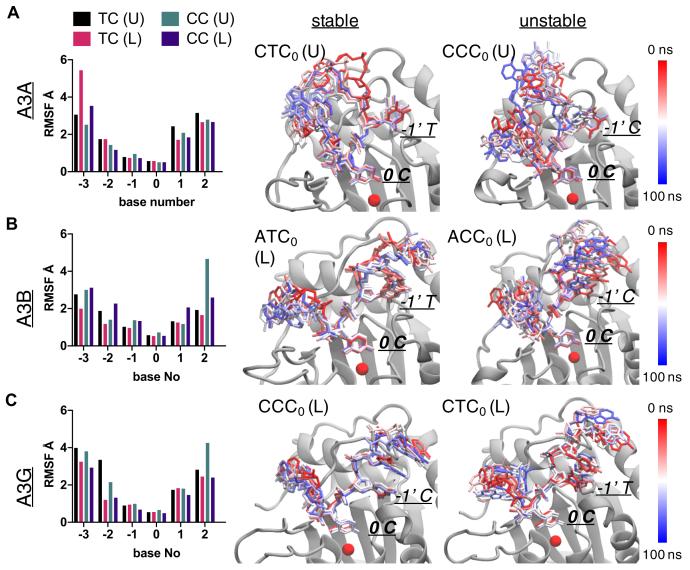
Figure 2.
Differential dynamics of ssDNA during the pMD simulations elucidate the stable substrate-bound A3 complexes. The root-mean-squared fluctuations (RMSF) of each nucleotide in four complexes with different sequences (TC or CC) and binding conformations (U or L) are shown as histograms. Two examples of stable (middle column) and unstable (right column) complexes for each of (A) A3A, (B) A3B-CTD, (C) A3G-CTD are depicted. In each panel, seven snapshots of the ssDNA conformation evenly spaced throughout a 100 ns MD trajectory are shown. The A3 proteins are in gray cartoon representation. The ssDNA is shown as stick; colored based on the simulation time from red to blue. The target (0 C) and −1′ position nucleotide in the active site are labeled. The catalytic zinc is shown as red sphere.