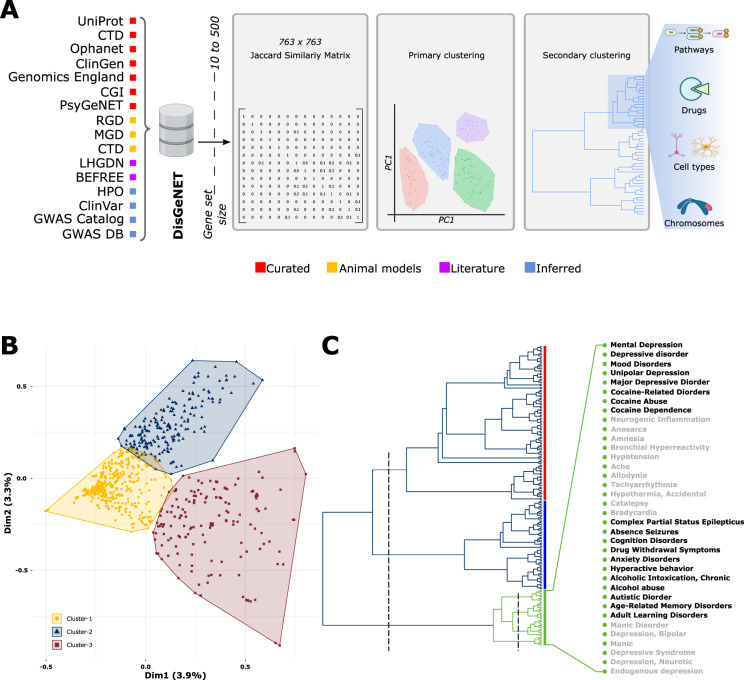
Fig. 1. Disease-associated gene-sets fall into three distinct clusters.
A Workflow used to derive the “psychiatric cluster” in an unbiased top-down manner. Curated gene-sets form different sources cataloged in DisGeNET’s were filtered by gene-set size, compared via a Jaccard similarity matrix and sorted into global clusters of similar disease using principal component analysis. Then, a primary cluster enriched in psychiatric, immune, metabolic, and neurodegenerative disorders was further broken down based on gene-set similarity, ultimately yielding the “psychiatric cluster” of 36 diseases that was used for subsequent analysis of pathways, drugs, cell-types, and chromosomes. B A PCA based clustering of 763 curated disorders. Note that cluster 1 falls at the center of the plot indicating a low internal correlation between the disorders in this cluster. C Hierarchical clustering of Cluster 2 disorders. An initial cutting of three (left dashed line) reveals three branches shown in red, blue, and green. The green branch (aka the psychiatric cluster) was considered for all subsequent analysis. Further cutting the green branch (right dashed line) reveals four subgroups composed of 36 disorders labeled on the right. See Supplementary Fig. 1 for further details. CTD Comparative Toxicogenomics Database, ClinGen The Clinical Genome Resource, CGI The Cancer Genome Interpreter, PsyGeNET Psychiatric disorders Gene association NETwork, RGD Rat Genome Database, MGD Mouse Genome Database, LHGDN Literature-derived Human Gene-Disease Network, HPO Human Phenotype Ontology, GWAS Gene-Wide Association Study.