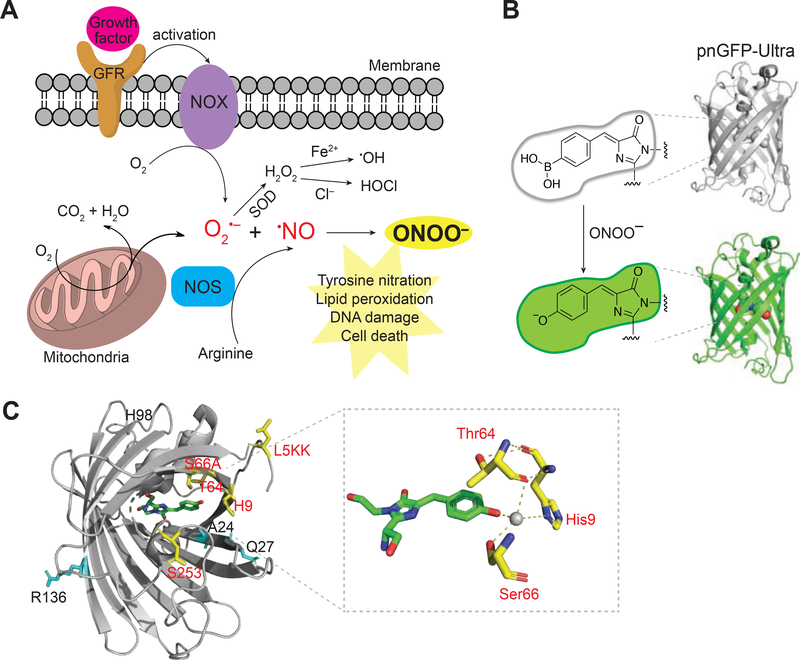
Figure 1. Peroxynitrite Biogenesis and Sensing Mechanism of pnGFP-Ultra.
(A) Illustration of the major peroxynitrite (ONOO–) biogenesis pathway. ONOO– is generated from the diffusion-controlled reaction between superoxide (O2•–) and nitric oxide (•NO). O2•– could be produced during mitochondrial respiration (oxidative phosphorylation) or by activated NADPH oxidase (NOX). NOX can be activated during cell signaling such as a growth factor binding to a growth factor receptor (GFR). Superoxide dismutase (SOD) can convert O2•– to hydrogen peroxide (H2O2), which can mediate secondary radical (e.g. •OH or –OCl) production. •NO is produced by nitric oxide synthetase (NOS), which is coupled to arginine metabolism. ONOO– is a highly reactive nitrogen species that plays crucial roles in tyrosine nitration, lipid peroxidation, DNA damage, and cell death.
(B) Sensing mechanism of pnGFP-Ultra. pnGFP-Ultra has a boronic acid-modified chromophore that can be converted to a phenolate chromophore upon reaction with ONOO–, resulting in a large fluorescence turn-on response.
(C) Cartoon representation of pnGFP1.5-Y.Cro (PDB 5F9G) and key residues targeted during pnGFP-Ultra development. Residues identified during directed evolution that increase the brightness and folding of cpsGFP are highlighted as cyan sticks. Residues that tune of the chemoselectivity of pnGFP-Ultra are highlighted as yellow sticks. The chromophore is show in green. The inset box on the right shows the chromophore and its interacting residues. The grey sphere denotes a water molecule. The structure was prepared using Pymol.