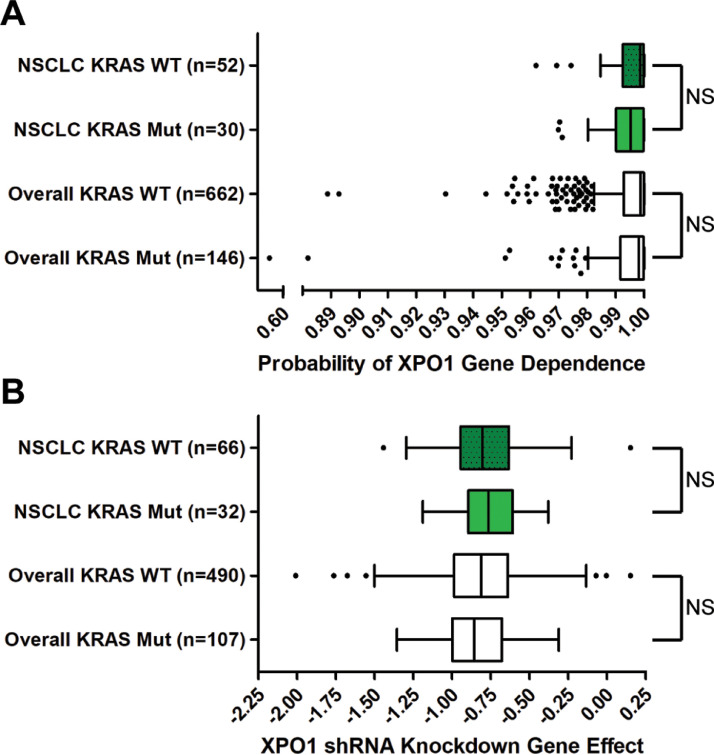
Fig. 5.
XPO1 gene dependency across different cancer cell lines. (A) Box and whisker plots of the probabilities of cancer cell line XPO1 genetic dependency from CRISPR/Cas9 knockout screens, compiled from the Achilles Project 20Q4 [37]. Values were estimated previously and shifted and scaled per cell line so a score of 0 represents 0% dependence on the gene of interest per cell line; a score of 1 represents 100% dependence on the gene of interest per cell line [37]. XPO1 dependency scores of KRASWT and KRASmut NSCLC cell lines (p = 0.27) or all cell lines overall (not restricted to NSCLC) (p = 0.066) were not significantly different. (B) Box and whisker plots of the probabilities of cancer cell line XPO1 genetic dependencies from RNAi knockdown screens, compiled from the Achilles Project [38]. Values were estimated previously, with a lower value representing a greater dependence on XPO1 for survival, although no scale was explicitly defined [38]. XPO1 dependency scores of KRASWT and KRASmut NSCLC cell lines (p = 0.58) or all cell lines overall (not restricted to NSCLC) (p = 0.43) were not significantly different. Mann-Whitney tests were used to compare means of each group. Tukey whiskers are plotted. NS = not significant.