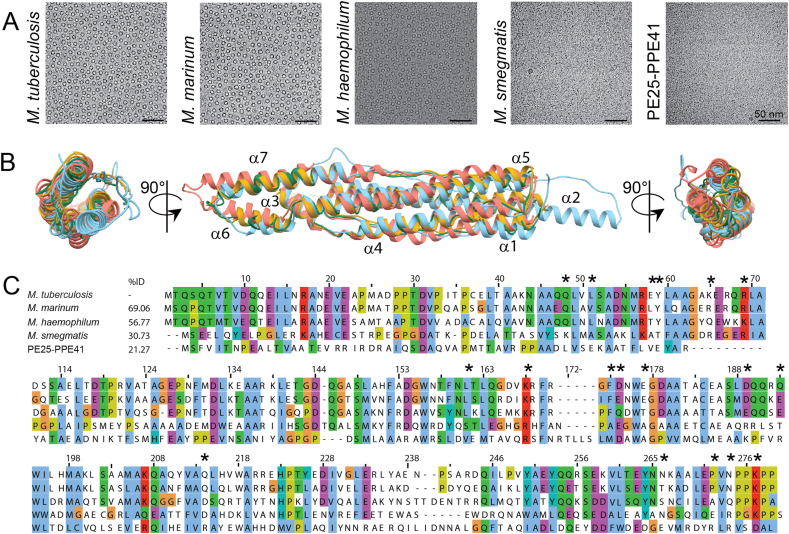
Fig. 3.
Oligomerisation differences between EspB orthologues despite sharing similar tertiary structure. (A) Evaluation of the oligomerisation of EspB orthologues and PE25-PPE41 by cryo-electron microscopy. Scale bars represent 50 nm. (B) Different views of structural alignment of EspBMtb (yellow – this work), EspBMmar (green – this work), EspBMsmeg (light blue – PDB ID 4WJ1), and PE25-PPE41 (orange – PDB ID 4W4K). (C) Multi-alignment of amino acid sequences of different species from the Mycobacterium genus, as well as the protein pair PE25-PPE41. Numbering and sequence identity is based on the sequence of M. tuberculosis. Asterisks denote residues making inter-subunit contact in EspB from M. tuberculosis and M. marinum. Alignment was generated using ClustalW server, and figure was created using software Jalview (Waterhouse et al., 2009). The colour scheme of ClustalX is used (Larkin et al., 2007).