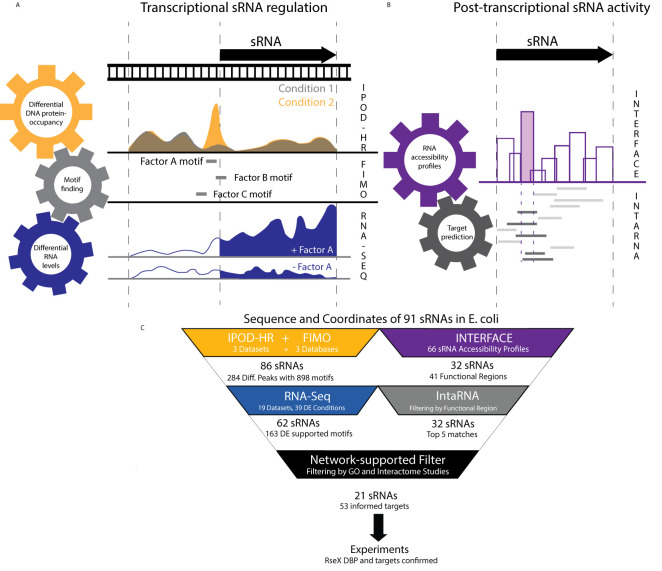
Figure 1.
An integrative top-down datamining approach utilizes publicly-available omics datasets to understand cellular regulation of and by any sRNA of interest. (A) The ID-sRnA approach is split into two distinct nodes. In the transcriptional regulation characterization node, DNA sequences corresponding to relevant sRNA transcription sequence space [-200 to +10] are narrowed to those that exhibit condition-specific occupancy. For selected 60-185 nt genomic fragments, DBP motif searching is performed to compile a set of putative regulators. High-confidence regulators are selected as DBPs for which differential RNA expression corroborates putative DBP binding. (B) The post-transcriptional sRNA characterization node of ID-sRnA relies on coupling of high-throughput regional accessibility data with computational target predictions. Region 3 (shaded) is selected as a likely functional region due to its toehold-like activity; namely, high accessibility with surrounding low accessibility. Target predictions are flagged by reliance of the lowest energy sRNA-mRNA interaction on the proposed sRNA functional region (dark) and re-ranked to exclude those that do not rely on the functional region for interaction. (C) Specific number of sRNAs and datasets used for each step of the ID-sRnA pipeline are highlighted, quantifying the amount of filtering performed at each step. 91 sRNAs were considered in which 62 and 32 sRNAs remain in the transcription and post-transcriptional regulation nodes, respectively. Fifty-three accessibility-informed targets corresponding to 21 sRNAs are supported by sRNA regulation factors identified through node 1 and/or previously documented sRNA characterization. Results for RseX are followed-up experimentally to confirm a negative DBP regulator (H-NS) as well as two novel targets, fimB and ihfB.