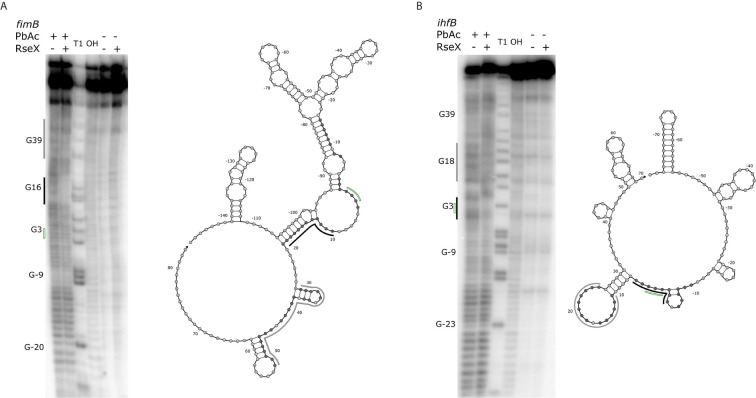
Figure 8.
Results of PbAc probing of representative 5’-labeled fimB (A) and ihfB (B) transcripts confirm RseX protection in IntaRNA-predicted regions. Position of various G residues (as concluded from guanine “T1” and alkaline “OH” ladders) are labeled to the left of the probing images, numbered with respect to the start codon (green, no fill). Control reactions without PbAc indicate initial levels of cleavage. By comparing the mRNA levels of cleavage with and without RseX, regions exhibiting strong (black) and weak (grey) protection were identified as interaction sites and are outlined to the left of the probing images. Nucleotides thermodynamically predicted to interact with RseX (shaded), start codon (green, no fill), as well as corresponding regions of strong and weak RseX protection (black, grey traces) are overlaid on corresponding Nupack-predicted secondary structures (Zadeh et al., 2011) of the mRNA 5’ UTRs through the predicted RseX interaction sites.