Figure 1: Identification of Pre-mRNA Splicing Components as Repressors of OXPHOS. See also Figure S1 and Table S1.
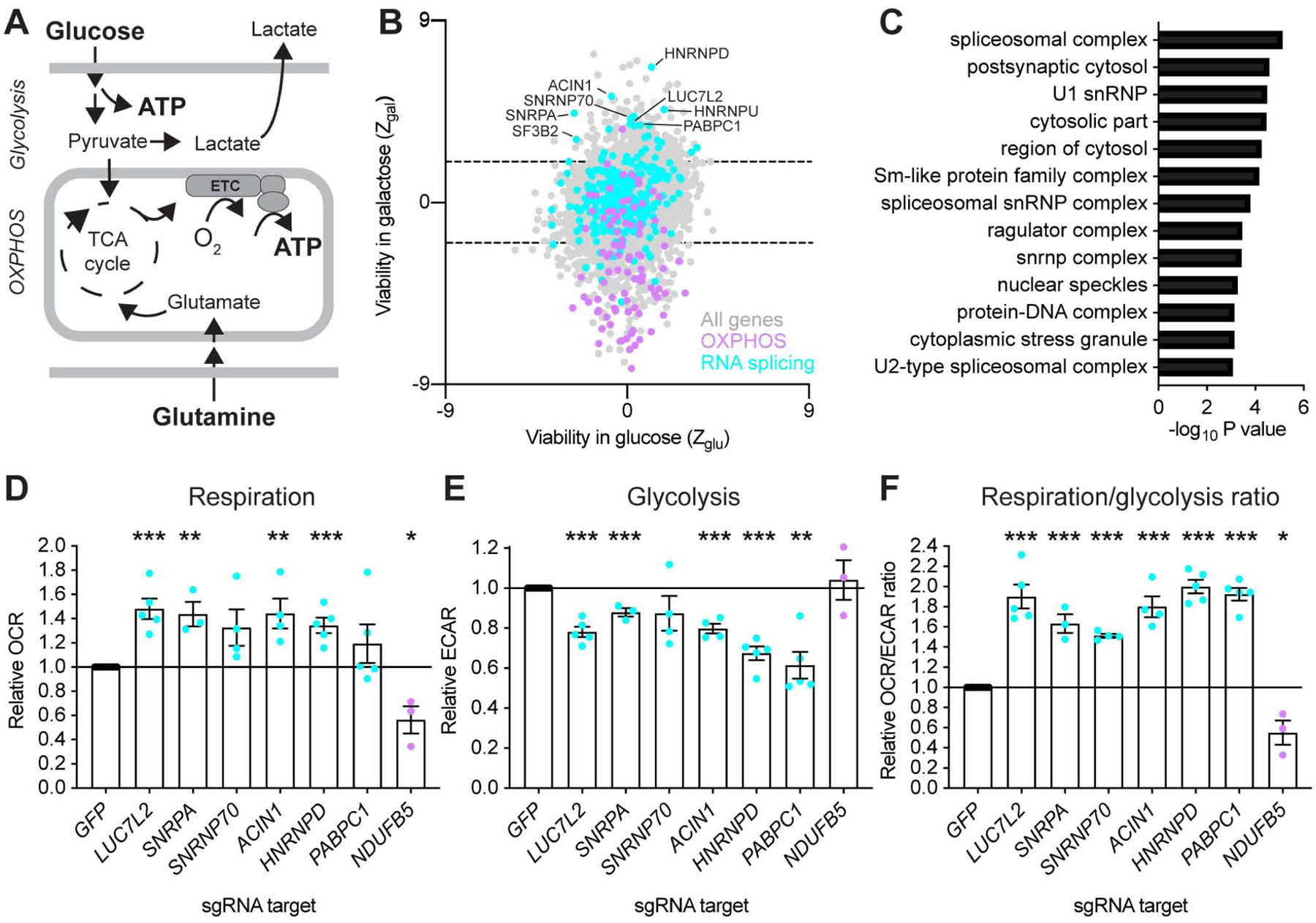
(A) Overview of the main ATP-generating pathways in human cells. OXPHOS: oxidative phosphorylation. ETC: electron transport chain. TCA: tricarboxylic acid cycle. (B) Gene-level analysis of a genome-wide CRISPR/Cas9 screen in glucose and galactose. Each dot represents an expressed, non-essential gene (n = 9,189). (C) Gene ontology analysis generated using a gene list ranked by viability in galactose against GO components. (D–F) Functional validation of the screening results. Basal whole-cell oxygen consumption rates (OCR), extracellular acidification rates (ECAR) and OCR/ECAR ratios were simultaneously measured after CRISPR/Cas9-mediated gene depletion in K562 cells grown in glucose-containing media. Data are shown as mean ± SEM (n≥3 independent experiments. *P<0.05, **P<0.01, ***P<0.001, t-test relative to control (GFP) sgRNA-treated cells. NDUFB5 is a control with known role in OXPHOS.
