Figure 5: Role of LUC7L2-Mediated PFKM and SLC7A11 Alternative Splicing in Energy Metabolism. See also Figure S5.
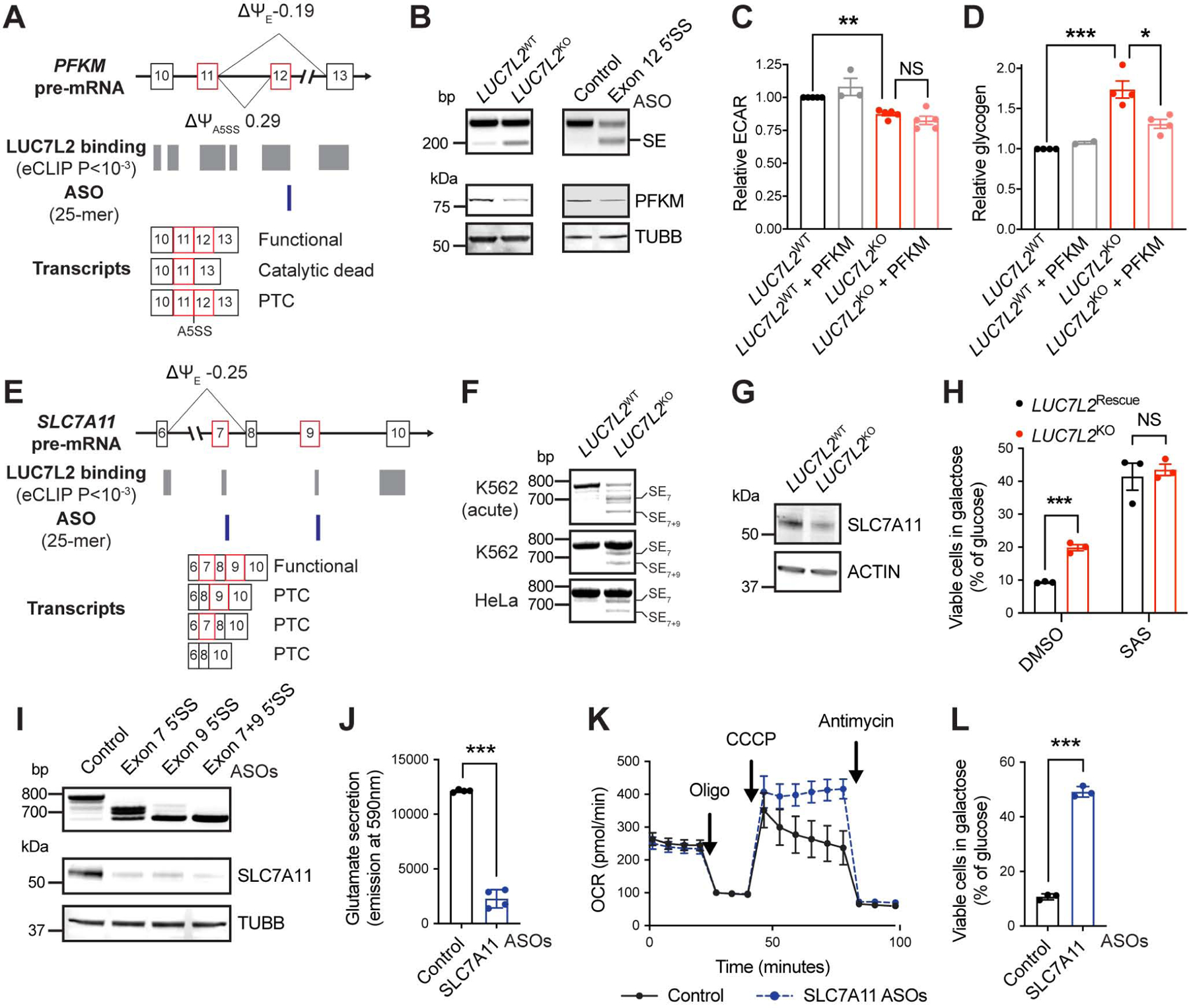
(A) Representation of PFKM exons 10–13, LUC7L2 binding sites as determined by eCLIP, antisense oligonucleotides (ASO) targeting sites and the expected transcripts. Ψ: percent spliced in reported by rMATS in K562 cells. E: exon. A negative Δψ value indicates exon skipping. PTC: premature termination codons. (B) RT-PCR (top) and immunoblot (bottom) of LUC7L2KO K562 cells (left) or HAP1 cells treated for 48h with ASO targeting the 5′SS of PFKM exon 12 (right). (C) Relative ECAR (n = 3–5) and (D) glycogen in LUC7L2KO K562 cells expressing control cDNAs (GFP) or PFKM cDNA (n = 2–4). (E) Representation of SLC7A11 exons 6–10 as in (A). (F) RT-PCR of LUC7L2KO K562 cells with primers amplifying transcripts corresponding to SLC7A11 exons 6–12. SE: skipped exon. (G) Immunoblot on LUC7L2KO K562 cells with antibodies to SLC7A11 and ACTIN. (H) Cell viability of LUC7L2Rescue (corresponds to LUC72L2KO expressing LUC7L2 cDNA) and LUC7L2KO HAP1 cells grown for 24h in galactose relative to glucose (n = 3). SAS: 500μM sulfasalazine. (I) RT-PCR (top) and immunoblot (bottom) of HAP1 cells treated for 48h with ASOs targeting the 5′SS of exon 7 and/or exon 9 of SLC7A11. (J) Media glutamate (n = 4), (K) representative seahorse trace (shown as mean ±SD), and (L) viability in galactose of HAP1 cells treated for 48h with the indicated ASOs (n = 3). All data are shown as mean ±SEM (unless otherwise stated) with * P<0.05, ** P<0.01, ***P<0.001, t-test relative to control.
