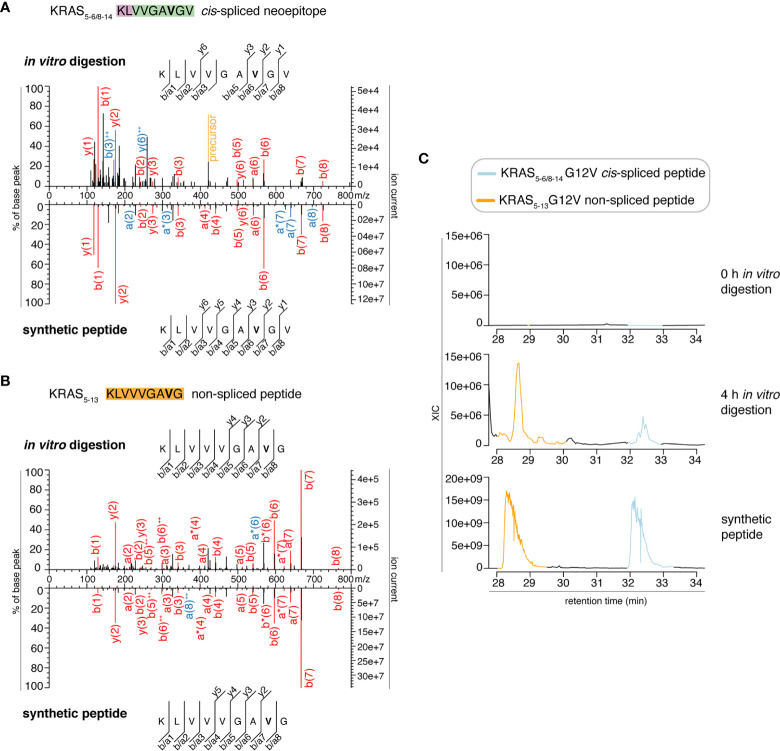
Figure 2.
MS2 spectra of the KRAS5-6/8-14 G12V cis-spliced and KRAS5-13 G12V non-spliced peptides by standard MS through Exploris 480 Orbitrap. (A, B) MS2 spectra of the KRAS5-6/8-14 G12V [KL][VVGAVGV] cis-spliced peptide (A) and KRAS5-13 G12V non-spliced peptide [KLVVVGAVG], which were identified in the 4 h in vitro digestions of the synthetic polypeptide KRAS2-35 G12V with purified proteasomes, in comparison with the cognate synthetic peptides. The peptide sequence is shown with the corresponding b-, a- and y-ions identified. The G12V mutation is depicted in bold. In the spectra, assigned peaks for b-, a- and y-ions are reported in color. Ion neutral loss of ammonia and double charged ions are symbolized by * and ++, respectively. Red marked peaks are assigned in MS2 spectra of both in vitro digestions and synthetic peptides, whereas blue marked peaks are assigned only in one of the two spectra. We marked the doubly charged peptide precursor in yellow. (C) Extracted ion chromatograms for the target peptides in the 0 h and 4 h in vitro digestion and the synthetic counterpart are plotted and indicate matching RTs and absence of a biologically significant peak in the 0 h digestion. MS ion chromatograms correspond to the m/z = 421.275 – 421.875. In (A–C) samples correspond to MF series carried out in HMDA buffer and have been measured through standard MS by Orbitrap Exploris 480 mass spectrometer.