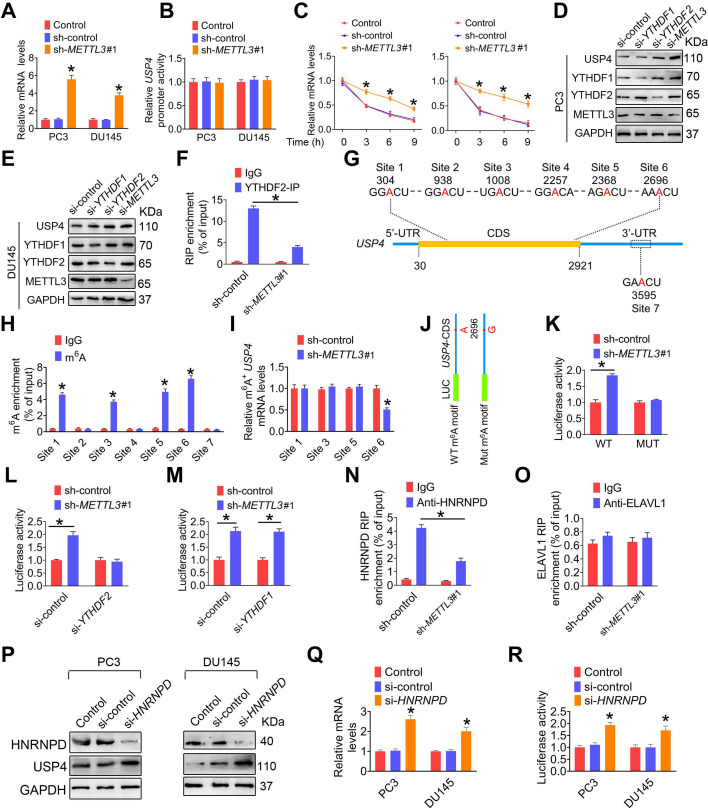
Figure 6.
m6A induces decay of the USP4 transcript in prostate cancer (PCa) cell. A-C, PC3 and DU145 cells were transfected with shRNA targeting METTL3 (sh-METTL3), respectively. The USP4 mRNA levels in indicated cells were determined by qRT-PCR assay (A). The USP4 promoter constructs were transfected into indicated cells, and luciferase activity was measured (B). PCa cells were treated with actinomycin D (5 μg/mL) for 2 h, followed by measurement of USP4 mRNA levels at indicated times (C). Data were presented as means ± SEM (n = 3), * p < 0.05 vs. the control cells. D-E, PC3 cells (D) and DU145 cells (E) were treated with indicated siRNAs, and cell lysates were subject to western blotting. F, Lysates from the PC3 cells were subjected to immunoprecipitation with anti-YTHDF2, and the association of the USP4 transcript with YTHDF2 was determined by qRT-PCR. Data were presented as means ± SEM (n = 3), * p < 0.05. G, Schematic representation of the position of m6A motifs within USP4 transcript. H, Abundance of USP4 transcript among mRNA immunoprecipitated with anti-m6A antibody was measured by qRT-PCR and normalized to input. Data are presented as means ± SEM (n = 3). * p < 0.05 vs. the IgG group. I, Abundance of USP4 transcript among mRNA immunoprecipitated with anti-m6A antibody was measured by qRT-PCR. Data are presented as means ± SEM (n = 3). * p < 0.05 vs. the control cells. J, USP4-CDS of the wild-type or mutant (A to G) was fused with a luciferase reporter. K-M, Luciferase activity of USP4-CDS was measured and normalized to Renilla luciferase activity. Data are presented as means ± SEM (n = 3). * p < 0.05. N-O, Lysates from the indicated cells were subjected to immunoprecipitation with anti-HNRNPD (N) or anti-ELAVL1 (O), and the association of the USP4 transcript with each protein was determined by qRT-PCR. Data were presented as means ± SEM (n = 3), * p < 0.05. P-R, PC3 cells and DU145 cells were treated with indicated siRNAs, and cell lysates were subject to western blotting (P). The USP4 mRNA levels in indicated cells were determined by qRT-PCR (Q). Luciferase activity of USP4-CDS in indicated cells was measured and normalized to Renilla luciferase activity (R). Data are presented as means ± SEM (n = 3), * p < 0.05 vs. the control cells.