Abstract
Background
Fluoroquinolone resistance in Mycobacterium tuberculosis (Mtb) is conferred by DNA gyrase mutations, but not all fluoroquinolone-resistant Mtb isolates have mutations detected. The optimal allele frequency threshold to identify resistance-conferring mutations by whole-genome sequencing is unknown.
Methods
Phenotypically ofloxacin-resistant and lineage-matched ofloxacin-susceptible Mtb isolates underwent whole-genome sequencing at an average coverage depth of 868 reads. Polymorphisms within the quinolone-resistance–determining region (QRDR) of gyrA and gyrB were identified. The allele frequency threshold using the Genome Analysis Toolkit pipeline was ~8%; allele-level data identified the predominant variant allele frequency and mutational burden (ie, sum of all variant allele frequencies in the QRDR) in gyrA, gyrB, and gyrA + gyrB for each isolate. Receiver operating characteristic (ROC) curves assessed the optimal measure of allele frequency and potential thresholds for identifying phenotypically resistant isolates.
Results
Of 42 ofloxacin-resistant Mtb isolates, area under the ROC curve (AUC) was highest for predominant variant allele frequency, so that measure was used to evaluate optimal mutation detection thresholds. AUCs for 8%, 2.5%, and 0.8% thresholds were 0.8452, 0.9286, and 0.9069, respectively. Sensitivity and specificity were 69% and 100% for 8%, 86% and 100% for 2.5%, 91% and 91% for 0.8%. The sensitivity of the 2.5% and 0.8% thresholds were significantly higher than the 8% threshold (P = .016 and .004, respectively) but not significantly different between one another (P = .5).
Conclusions
A predominant mutation allele frequency threshold of 2.5% had the highest AUC for detecting DNA gyrase mutations that confer ofloxacin resistance, and was therefore the optimal threshold.
Keywords: M. tuberculosis, fluoroquinolone resistance, whole-genome sequencing
Not all fluoroquinolone-resistant M. tuberculosis isolates have mutations detected in DNA gyrase. The optimal allele frequency threshold to identify resistance-conferring mutations by whole-genome sequencing is unknown. In this study, an allele frequency threshold of 2.5% had optimal sensitivity and specificity.
Fluoroquinolones have potent bactericidal activity against Mycobacterium tuberculosis; they act by binding to DNA gyrase and preventing negative supercoiling of replicating DNA. Mutations in gyrA and gyrB, the genes that encode the 2 subunits of DNA gyrase, are the most frequent mechanism of fluoroquinolone resistance [1, 2]. Such mutations confer resistance by interfering with fluoroquinolones binding to M. tuberculosis in the quinolone binding pocket (QBP), within the quinolone-resistance–determining region (QRDR). The QRDR is composed of codons 74–113 in gyrA and 461–499 in gyrB (using the P0C5C5|1–675 numbering scheme) [3]. Recent studies have identified fluoroquinolone-resistance–conferring mutations immediately outside the traditional QRDR of gyrB, at codons 500 and 501. These 2 codons are in the QBP and in direct contact with the quinolone molecule. These studies refined the definition of the QRDR in gyrB, extending it from position 461 to 501 [1, 4].
Whole-genome sequencing is a sensitive technique for identifying resistance mutations. However, not all fluoroquinolone-resistant M. tuberculosis isolates have mutations identified in DNA gyrase [2, 5–7]. Although web-based tools have been developed to improve resistance-conferring mutation identification, a consensus regarding the depth of coverage required for whole-genome sequencing to detect drug resistance in M. tuberculosis has not been established [6, 8].
In a previous study, we noted heteroresistance in 10 of 26 (38%) M. tuberculosis isolates phenotypically resistant to ofloxacin. Allele frequencies ranging from 4% to 84% were observed in the heteroresistant isolates [9].
Although diagnostic tests accurately detect resistance to first-line antituberculosis (-TB) drugs such as isoniazid and rifampin, more sensitive tests are needed to detect fluoroquinolone resistance [10, 11]. A recent study using targeted next-generation sequencing found that microheteroresistance exists below the resolving capability of conventional genotypic tests [12].
In this study, we sought to extend our previous findings by reporting on the allele-level frequency of variants seen in fluoroquinolone-susceptible and -resistant M. tuberculosis isolates. We investigated whether a widely used genome-wide bioinformatics pipeline missed low-level fluoroquinolone-resistant variants (ie, microheteroresistance), and propose using allele-level analysis at an allele frequency threshold that optimizes sensitivity and specificity of genotypic fluoroquinolone resistance, to complement whole-genome analysis.
METHODS
Study Overview
We performed whole-genome sequencing and assessed allele-level data for DNA gyrase variants in 3 groups of M. tuberculosis isolates: (1) phenotypically ofloxacin-resistant isolates with gyrA/B mutations identified on whole-genome sequencing, (2) ofloxacin-resistant isolates without gyrA/B mutations identified on whole-genome sequencing, and (3) ofloxacin-susceptible isolates.
Study Population
Ofloxacin-resistant M. tuberculosis isolates were identified in a population-based study of patients with newly diagnosed, culture-confirmed TB reported in Tennessee from 2002 to 2016. Eleven phenotypically ofloxacin-susceptible isolates were selected on the basis of genotypic lineage, which was determined by spoligotype and 12-locus mycobacterial interspersed repetitive-unit (MIRU) compared with 13 ofloxacin-resistant isolates that did not have gyrA/B mutations by whole-genome sequencing [13]. Controls for these latter isolates were felt to be most important, since they were felt likely to have an alternative fluoroquinolone-resistance mechanism. One fluoroquinolone-resistant isolate of Indo-Oceanic lineage did not have any matched fluoroquinolone-susceptible isolates.
Resistance Testing
All isolates in this study were tested at Vanderbilt University Medical Center (VUMC) by 2 different drug-susceptibility methods: agar proportion and the resazurin microtiter assay (REMA). Briefly, a suspension with a turbidity equivalent to a 1.0 McFarland standard was prepared from <14-day-old M. tuberculosis colonies grown on Lowenstein–Jensen medium. After confirmation of turbidity by nephelometer, the suspension served as the standard inoculum for all dilutions. One hundred microliters each of 10−2 and 10−4 dilutions of the standard inoculum were plated on 7H10 agar with and without ofloxacin. Phenotypic resistance was defined as more than 1% colony growth in the presence of 2 mg/L ofloxacin compared with colony growth in the absence of drug.
REMA has been validated for ofloxacin resistance testing in M. tuberculosis, using a critical concentration of 2 mg/L [9, 14, 15]. The ofloxacin (Sigma-Aldrich, St Louis, MO) minimum inhibitory concentration (MIC) was assessed at 2-fold dilutions between 0.5 and 64 mg/L. If growth was observed at day 7 in the control well, 30 μL of 0.01% resazurin solution was added to each well, followed by 24 hours of incubation. Color change indicated bacterial growth and MIC was determined as the lowest ofloxacin concentration preventing color change. The critical concentration of ofloxacin was 2 mg/L. The MIC reported and used for analyses was the median of 3 tests performed (Supplementary Table 1).
Molecular Typing
Spoligotyping and 12-locus MIRU typing were performed by the Michigan Department of Community Health on all M. tuberculosis isolates from culture-confirmed TB cases in Tennessee from 2004 to 2016 and a select number of isolates from 2002 and 2003 [13].
Whole-Genome Sequencing
Each isolate was grown on Lowenstein-Jensen media in the absence of ofloxacin. Multiple colonies from each isolate were emulsified in 200 µL nuclease-free water. Genomic DNA was isolated from the cell suspension and purified using the ZR Bacterial/Fungal DNA Mini-prep Kit (Zymo Research, Irvine, CA). Samples were then heated to 100oC for 10 minutes. The DNA concentration was verified by NanoDrop (Thermo Scientific, Waltham, MA).
Whole-genome sequencing was performed at the Genome Sciences Resource at VUMC for 53 M. tuberculosis clinical isolates (42 phenotypically ofloxacin resistant, 11 susceptible). The Illumina HiSEq, HiSEq 3000, and Novaseq platforms generated paired-end reads of 100 and 150 bp with an average depth of coverage of 868 reads (Supplementary Table 2). Raw base calls were generated from the internal Casava pipeline software (Illumina, San Diego, CA). Output was in the fastq format. Sequences were aligned and mapped to the reference H37Rv genome (Genbank Al123456.3) using the Burrows Wheeler Aligner [16]. The alignments were sorted and indexed using SAMtools [17]. The unified genotyper from the Genome Analysis Toolkit (GATK) was then used to make variant calls [18]. GATK output contained an unfiltered list of single nucleotide polymorphisms (SNPs) in the variant call format. SNPeff was used to annotate output base changes and determine whether mutations were synonymous [19].
Allele-level Analysis
For each sample, allele frequencies of the 4 nucleotides at 27 selected loci in gyrA and gyrB were computed from pileup results of Binary Alignment Map (BAM) files, by counting reads aligned to each nucleotide. Sequencing data went through rigorous quality-control procedures. Sequence reads with a mapping quality Phred score less than 20 (reads with error rate >1%) were removed from the analysis. Low-quality reads were filtered out before allele frequencies were counted [20].
Genomic positions were selected from codons in the QRDRs of gyrA/B that were confirmed to confer fluoroquinolone resistance via functional analysis; this was determined by review of the literature and available databases (ReSeqTB) [2, 4–7, 21–33] (Supplementary Table 3).
Statistical Methods
The predominant DNA gyrase mutation was defined as the variant with the highest nucleotide allele frequency among the selected loci in gyrA/B. The mutational burden in gyrA was determined by summing all variant allele frequencies in the gyrA loci. The same process was performed among the selected loci in gyrB to determine the mutational burden in gyrB. The total mutational burden was the sum of variant allele frequencies in gyrA/B.
The Kruskal-Wallis and Wilcoxon rank-sum tests assessed differences in the distribution of predominant allele frequency and mutational burdens among resistant and susceptible isolates.
To identify the measure of variant allele frequency that correlated most closely with phenotypic fluoroquinolone resistance, we performed logistic regression of the binary phenotypic resistance outcome on the continuous allele frequency exposure to generate a receiver operating characteristic (ROC) curve and calculated the area under the curve (AUC). These analyses are used to assess the discrimination of a binary outcome (eg, resistant vs not resistant) using predictors (eg, allele frequency), and to find the optimal sensitivity and specificity threshold values of a test and compare these values across more than 1 diagnostic test [34, 35]. The ROC of a test with perfect discrimination passes through the left-upper corner of the unit square (the point where the sensitivity and specificity are equal to 1) [36]. Each observation generates a binary response classification matrix in the form of a predicted probability of the positive or negative result. A threshold on the predictive probability scale can be selected, above which the test is positive. Considering phenotypic drug-susceptibility testing as the reference standard, we compared it with (1) the mutation in gyrA or gyrB with the predominant allele frequency, (2) the mutational burden in gyrA, (3) the mutational burden in gyrB, and (4) the total mutational burden.
Subsequently, to identify the optimal allele frequency cutoff (genotypic threshold) for detecting phenotypic fluoroquinolone resistance, we used 3 commonly used optimality criteria for determining threshold values: the point on the ROC curve (1) with minimum distance from the left-upper corner of the unit square, (2) where the sensitivity and specificity of the test were equal, and (3) where the sensitivity and specificity were maximized (Youden’s index). These 3 criteria weight sensitivity and specificity equally [36].
Once the optimal threshold values were determined, we assessed the allele frequency detection threshold of whole-genome sequencing analysis and considered the lowest allele frequency that made a variant call in the genome-wide analysis as the threshold. We calculated the sensitivity and specificity when using each of the optimal thresholds and compared them with the genome-wide analysis threshold using the McNemar’s test to determine statistical significance. SAS system for Windows version 7.15 HF7 (SAS Institute, Cary, NC) was used for all analyses.
RESULTS
There were 42 ofloxacin-resistant M. tuberculosis isolates during the study period, of which 29 (69%) had resistance-conferring mutations in gyrA or gyrB on whole-genome sequencing and 13 (31%) did not. There were 11 ofloxacin-susceptible M. tuberculosis isolates. Output files were combined for all 53 isolates. After removing SNPs found in susceptible isolates and synonymous SNPs, 19 unique nonsynonymous SNPs were identified in gyrA or gyrB on whole-genome sequencing; 11 of 19 were inside the QRDR of gyrA or gyrB.
Table 1 shows all nonsynonymous mutations in the QRDRs of gyrA/B found in the study isolates. Five of the mutations have previously been shown to confer resistance by functional analysis; the remaining mutations have been reported in fluoroquinolone-resistant isolates [2, 25]. Mutations at positions 7582(D94G) and 7581(D94N) were the most frequent mutations in gyrA, found in 13 and 7 ofloxacin-resistant isolates, respectively. The A90V mutation at position 7570 was seen in 6 ofloxacin-resistant isolates and the mutations at positions 7572(S91P) and 7581(D94Y) were each seen in 3 ofloxacin-resistant isolates. The D94A and D89N at positions 7582 and 7566 were each seen in 1 ofloxacin-resistant isolate.
Table 1.
All Nonsynonymous Mutations at Codons in DNA Gyrase That Have Been Associated With Fluoroquinolone Resistance in Mycobacterium tuberculosis Identified in Our Study Population of M. tuberculosis Isolates
| Name | Gene | Position | AA_Change | Codon Changea |
|---|---|---|---|---|
| gyrA | Rv0006 | 7566 | D89N | Gac/Aac |
| gyrA | Rv0006 | 7570 | A90Vb | gCg/gTg |
| gyrA | Rv0006 | 7572 | S91P | Tcg/Ccg |
| gyrA | Rv0006 | 7581 | D94N | Gac/Aac |
| gyrA | Rv0006 | 7581 | D94Y | Gac/Tac |
| gyrA | Rv0006 | 7582 | D94A | gAc/gCc |
| gyrA | Rv0006 | 7582 | D94Gb | gAc/gGc |
| gyrB | Rv0005 | 6620 | D461Nb | Gac/Aac |
| gyrB | Rv0005 | 6620 | D461Hb | Gac/Cac |
| gyrB | Rv0005 | 6735 | N499I | aAc/aTc |
| gyrB | Rv0005 | 6738 | T500Nb | aCc/aAc |
The codon, not necessarily the mutation, has been associated with fluoroquinolone resistance.
Abbreviation: AA, amino acid.
aCapital letters denote nucleotide change.
bFluoroquinolone resistance mutations previously shown to confer resistance by functional analysis.
Four mutations were found in gyrB at codons 461(D461N and D461H), 499(N499I), and 500(T500N), each appearing in 1 ofloxacin-resistant isolate. Mutations at codons 461(D461H) and 500 were seen in isolates that also had gyrA mutations.
Of the 11 ofloxacin-susceptible isolates, none had resistance-conferring mutations in the QRDRs of gyrA/B.
Allele-level Analysis
Allele frequencies were obtained for 27 selected genomic positions (15 in gyrA and 12 in gyrB); 23 genomic positions remained after removing the positions resulting in synonymous substitutions (12 in gyrA and 11 in gyrB). Table 2 shows the allele frequency of the predominant DNA gyrase mutation and the gyrA, gyrB, and total mutational burdens for the 29 fluoroquinolone-resistant isolates with DNA gyrase mutations identified in the genome-wide analysis. The lowest allele frequency seen was 8%. Nucleotides with lower allele frequencies were not called variants in the genome-wide analysis. Table 3 shows the predominant allele frequency and the gyrA, gyrB, and total mutational burdens for the 13 fluoroquinolone-resistant isolates without DNA gyrase mutations by whole-genome sequencing analysis. The highest allele frequency was 6.8% and the lowest was 0.2%; 1 isolate had no allele frequencies greater than 0. Supplementary Table 4 includes all fluoroquinolone-resistant isolates from Tables 2 and 3, listed according to predominant allele frequency.
Table 2.
DNA Gyrase Allele Frequencies of Fluoroquinolone-resistant Mycobacterium tuberculosis Isolates With Mutations Identified on Whole-Genome Sequencing at Codons Known to Confer Fluoroquinolone Resistance
| Mutational Burdena | ||||||
|---|---|---|---|---|---|---|
| Study ID | MIC | Predominant DNA Gyrase Mutation Allele Frequency | gyrA | gyrB | gyrA Plus gyrB | Predominant DNA Gyrase Mutation |
| 1 | 8 | 1.0000 | 1.0110 | 0.0059 | 1.0169 | D94G |
| 2 | 8 | 1.0000 | 1.0097 | 0.0027 | 1.0125 | D94Y |
| 3 | 8 | 1.0000 | 1.0082 | 0.0097 | 1.0180 | D94G |
| 4 | 16 | 0.9992 | 1.0177 | 0.0117 | 1.0295 | D94G |
| 5 | 32 | 0.9989 | 1.0157 | 0.0156 | 1.0313 | D94G |
| 6 | 16 | 0.9988 | 1.0178 | 0.0123 | 1.0301 | S91P |
| 7 | 4 | 0.9985 | 1.0217 | 0.0159 | 1.0376 | A90V |
| 8 | 16 | 0.9980 | 1.0095 | 0.0057 | 1.0151 | D94G |
| 9 | 16 | 0.9976 | 1.0110 | 0.0067 | 1.0177 | D94Y |
| 10 | 16 | 0.9976 | 1.0085 | 0.0123 | 1.0209 | D94N |
| 11 | 16 | 0.9975 | 0.9987 | 0.0033 | 1.0020 | D94N |
| 12 | 16 | 0.9960 | 1.0156 | 0.0096 | 1.0252 | D94G |
| 13 | 16 | 0.9772 | 1.0185 | 0.0082 | 1.0268 | D94N |
| 14 | 16 | 0.9686 | 0.9988 | 0.0091 | 1.0079 | D94N |
| 15 | 16 | 0.9621 | 0.9918 | 0.0260 | 1.0179 | D94G |
| 16 | 4 | 0.9583 | 0.0000 | 0.9583 | 0.9583 | N499I |
| 17 | 8 | 0.8098 | 0.8210 | 0.2067 | 1.0276 | A90V |
| 18 | 8 | 0.8060 | 1.0059 | 0.0133 | 1.0192 | D94N |
| 19 | 16 | 0.7983 | 0.9707 | 0.0035 | 0.9742 | D94Y |
| 20 | 16 | 0.6322 | 1.0161 | 0.0068 | 1.0230 | A90V |
| 21 | 8 | 0.4946 | 0.9958 | 0.0106 | 1.0064 | D94N |
| 22 | 16 | 0.4775 | 0.4841 | 0.0015 | 0.4856 | D94G |
| 23 | 16 | 0.4393 | 0.7420 | 0.2193 | 0.9613 | D94G |
| 24 | 8 | 0.3491 | 0.3491 | 0.0013 | 0.3504 | D94G |
| 25 | 4 | 0.2611 | 0.4706 | 0.0013 | 0.4719 | D94N |
| 26 | 8 | 0.2256 | 0.4609 | 0.0050 | 0.4659 | S91P |
| 27 | 4 | 0.0987 | 0.0110 | 0.1044 | 0.1153 | D461N |
| 28 | 4 | 0.0890 | 0.0930 | 0.0038 | 0.0968 | D94A |
| 29 | 4 | 0.0802b | 0.0938 | 0.0000 | 0.0938 | A90V |
N = 29.
Abbreviation: MIC, minimum inhibitory concentration.
aMutational burden: sum of mutation allele frequencies at resistance-conferring codons in the quinolone-resistance determining region (QRDR) of gyrA, gyrB, or both combined.
bMutation detection threshold by genome-wide analysis.
Table 3.
DNA Gyrase Allele Frequencies of Fluoroquinolone-resistant Mycobacterium tuberculosis Isolates Without Mutations Identified on Whole-Genome Sequencing at Codons Known to Confer Fluoroquinolone Resistance
| Mutational Burdena | ||||||
|---|---|---|---|---|---|---|
| Study ID | MIC | Predominant DNA Gyrase Mutation Allele Frequency | gyrA | gyrB | gyrA Plus gyrB | Predominant DNA Gyrase Mutation |
| 30 | 4 | 0.0675 | 0.0734 | 0.0018 | 0.0752 | D94G |
| 31 | 8 | 0.0594 | 0.0741 | 0.0086 | 0.0826 | D94H |
| 32 | 8 | 0.0481 | 0.0520 | 0.0010 | 0.0531 | D94N |
| 33 | 8 | 0.0290 | 0.0386 | 0.0000 | 0.0386 | D94Y |
| 34 | 2 | 0.0273 | 0.0371 | 0.0092 | 0.0463 | D94G |
| 35 | 8 | 0.0261 | 0.0348 | 0.0080 | 0.0429 | D94G |
| 36 | 8 | 0.0253 | 0.0323 | 0.0053 | 0.0376 | D94N |
| 37 | 8 | 0.0092 | 0.0223 | 0.0142 | 0.0365 | D89Y |
| 38 | 4 | 0.0075 | 0.0052 | 0.0075 | 0.0127 | D461Y |
| 39 | 8 | 0.0054 | 0.0143 | 0.0051 | 0.0193 | D94N |
| 40 | 8 | 0.0041 | 0.0164 | 0.0041 | 0.0205 | D461Y |
| 41 | 16 | 0.0021 | 0.0077 | 0.0038 | 0.0115 | A90T |
| 42b | 4 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | … |
N = 13.
Abbreviation: MIC, minimum inhibitory concentration.
aMutational burden: sum of variant allele frequencies at resistance-conferring codons in the quinolone-resistance determining region (QRDR) of gyrA, gyrB, or both combined.
bIn this isolate, all allele frequencies at the selected positions were 0.
In Table 4 the highest predominant allele frequency of the 11 fluoroquinolone-susceptible isolates was 1.7% and the lowest was 0.1%. We compared variant allele frequencies between the 3 groups of isolates. The differences in median allele frequencies between fluoroquinolone-resistant isolates with DNA gyrase mutations, without DNA gyrase mutations, and fluoroquinolone-susceptible isolates were all statistically significant for the predominant allele frequency, the mutational burden in gyrA, and the total mutational burden. However, the differences were not statistically significant for the gyrB mutational burden (Table 5).
Table 4.
DNA Gyrase Allele Frequencies of Fluoroquinolone-Susceptible Mycobacterium tuberculosis Isolates
| Mutational Burdena | ||||||
|---|---|---|---|---|---|---|
| Study ID | MIC | Predominant DNA Gyrase Mutation Allele Frequency | gyrA | gyrB | gyrA Plus gyrB | Predominant DNA Gyrase Mutation |
| 43 | 1 | 0.0173 | 0.0478 | 0.0142 | 0.0619 | D89Y |
| 44 | 1 | 0.0063 | 0.0255 | 0.0056 | 0.0311 | D94Y |
| 45 | 1 | 0.0060 | 0.0201 | 0.0122 | 0.0323 | D94Y |
| 46 | 1 | 0.0028 | 0.0122 | 0.0060 | 0.0182 | D461Y |
| 47 | 1 | 0.0025 | 0.0038 | 0.0049 | 0.0087 | G88S |
| 48 | 1 | 0.0025 | 0.0046 | 0.0025 | 0.0071 | D461Y |
| 49 | 1 | 0.0019 | 0.0019 | 0.0024 | 0.0043 | S91W |
| 50 | 1 | 0.0018 | 0.0020 | 0.0026 | 0.0047 | D461H |
| 51 | 1 | 0.0016 | 0.0025 | 0.0030 | 0.0054 | D94N |
| 52 | 1 | 0.0015 | 0.0043 | 0.0025 | 0.0068 | A90V |
| 53 | 1 | 0.0013 | 0.0026 | 0.0033 | 0.0059 | A90G |
None of these isolates had mutations identified on whole-genome sequencing at codons known to be associated with fluoroquinolone resistance (N = 11).
Abbreviation: MIC, minimum inhibitory concentration.
aMutational burden: sum of variant allele frequencies at resistance-conferring codons in the quinolone-resistance determining region (QRDR) of gyrA, gyrB, or both combined.
Table 5.
Differences in Median Mutation/Variant Allele Frequencies Among Fluoroquinolone-resistant Mycobacterium tuberculosis Isolates With and Without DNA Gyrase Mutations on Whole-Genome Sequencing and Fluoroquinolone-Susceptible Isolates
| Isolate Groupa | N | Predominant DNA Gyrase Mutation/ Variant Allele Frequency (Median) | P | Mutational Burden gyrA (Median) | P | Mutational Burden gyrB (Median) | P | Mutational Burden gyrA Plus gyrB (Median) | P |
|---|---|---|---|---|---|---|---|---|---|
| FQR with gyr mutations | 29 | 0.9621 | <.0001 | 0.9988 | <.0001 | 0.0091 | .0519 | 1.0151 | <.0001 |
| FQR without gyr mutations | 13 | 0.0253 | 0.0323 | 0.0051 | 0.0376 | ||||
| FQS | 11 | 0.0025 | 0.0043 | 0.0033 | 0.0071 | ||||
| FQR with gyr mutations | 29 | 0.9621 | <.0001 | 0.9988 | <.0001 | 0.0091 | .0517 | 1.0151 | <.0001 |
| FQR without gyr mutations | 13 | 0.0253 | 0.0323 | 0.0051 | 0.0376 | ||||
| FQR with gyr mutations | 29 | 0.9621 | <.0001 | 0.9988 | <.0001 | 0.0091 | .0579 | 1.0151 | <.0001 |
| FQS | 11 | 0.0025 | 0.0043 | 0.0033 | 0.0071 | ||||
| FQR without gyr mutations | 13 | 0.0253 | .0130 | 0.0323 | .0279 | 0.0051 | .9315 | 0.0376 | .0279 |
| FQS | 11 | 0.0025 | 0.0043 | 0.0033 | 0.0071 |
Abbreviations: FQR, fluoroquinolone-resistant; FQS, fluoroquinolone-susceptible.
aComparisons using 3 levels done by Kruskal-Wallis test; pairwise comparisons done by Wilcoxon rank-sum test.
Receiver Operating Characteristic Analyses
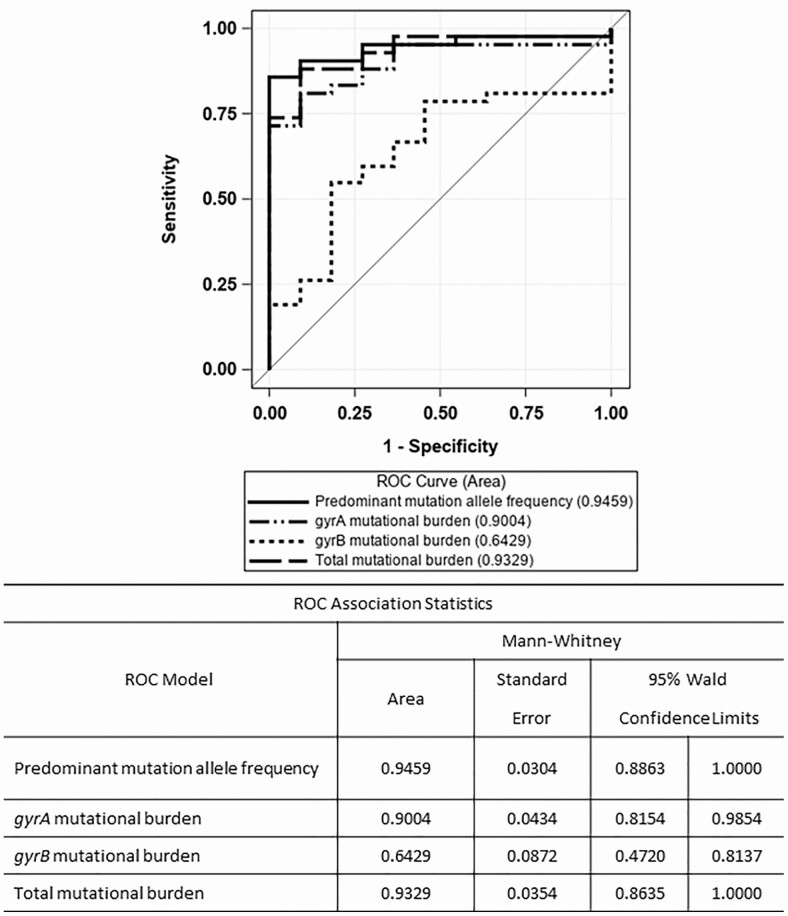
Figure 1 shows the ROC curve comparison to determine which measure (ie, the predominant allele frequency, gyrA mutational burden, gyrB mutational burden, or total DNA gyrase mutational burden) had the best predictive accuracy, using phenotypic drug susceptibility testing (DST) as the reference standard. The AUC of the predominant mutation allele frequency was highest at 0.95, and the gyrB mutational burden was the lowest at 0.64. The confidence interval for gyrB mutational burden (Figure 1) contains .5; hence, it is not significantly different from a random guess, which is represented by the diagonal line. The AUC is used as an index of discriminating ability; therefore, the measure with the highest AUC (ie, predominant mutation allele frequency) was used as the measure to predict fluoroquinolone resistance.
Figure 1.
ROC plot comparing phenotypic DST result with the proportion of reads associated with fluoroquinolone resistance in Mycobacterium tuberculosis DNA gyrase: (1) the mutation in gyrA or gyrB with the predominant allele frequency, (2) the sum of variant allele frequencies in gyrA (gyrA mutational burden), (3) the sum of variant allele frequencies in gyrB (gyrB mutational burden), or (4) the sum of variant allele frequencies in gyrA plus gyrB (total mutational burden). Abbreviations: DST, drug susceptibility test; ROC, receiver operating characteristic.
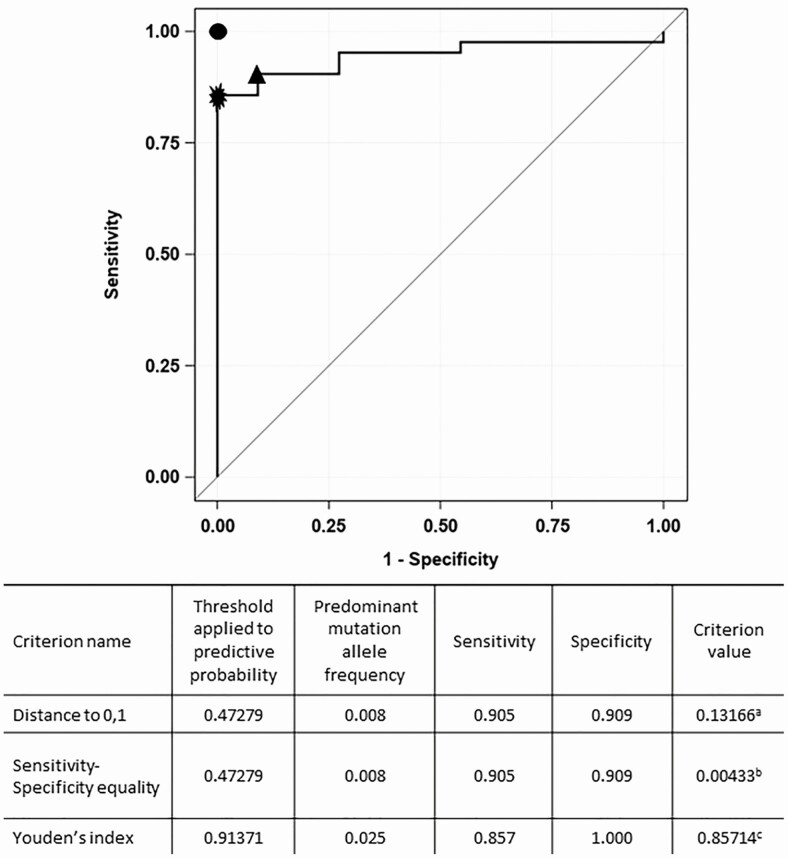
To analyze the effectiveness of the predominant mutation allele frequency as a predictor of fluoroquinolone resistance, we performed a second ROC curve analysis to identify optimal thresholds on the ROC curve using the 3 optimality criteria described above. Figure 2 shows the ROC curve with the optimal thresholds identified corresponding to the predominant allele frequency of 0.025 (2.5%) by 1 of the criteria used: Youden’s index. The second and third criteria—the minimum distance from the left-upper corner to the unit square and the point on the ROC curve where the sensitivity and specificity are equal—show a threshold corresponding to a predominant allele frequency of 0.008 (0.8%). The sensitivity and specificity of the 0.008 threshold was 91%, while the sensitivity and specificity of the 0.025 threshold were 86% and 100%, respectively (Figure 2, Table 6, Supplementary Tables 5 and 6).
Figure 2.
ROC plot comparing phenotypic DST with the predominant mutation allele frequency in gyrA or gyrB associated with fluoroquinolone resistance in Mycobacterium tuberculosis to identify the optimal genotypic threshold for detecting phenotypic resistance. A threshold of >2.5% was associated with the best sensitivity and specificity for detecting phenotypic fluoroquinolone resistance. The point at the left-upper corner shows where sensitivity = 1 and specificity = 1 or 100% (perfect test) is denoted by the filled circle. The optimal threshold corresponding to 0.025 predominant mutation allele frequency (denoted by the filled star) identified by Youden’s index criterion (the point on the ROC curve where sensitivity + specificity – 1 is maximum). The optimal threshold corresponding to 0.008 predominant allele frequency (denoted by the filled triangle) identified by distance to 0, 1 criterion (the point on the ROC curve where the distance to the perfect test is minimum). aCriterion value derived from the distance to the perfect point in the unit square [√ ([1 − sensitivity] [2] + [1 − specificity] [2]) = 0.131]. bCriterion value derived from the difference between sensitivity and specificity (sensitivity – specificity = 0.004). cCriterion value derived from the Youden’s index (sensitivity + specificity − 1 = 0.857). Abbreviations: DST, drug susceptibility test; ROC, receiver operating characteristic.
Table 6.
Sensitivity and Specificity of Different Thresholds of Whole-Genome Sequencing Compared to Phenotypic Drug Susceptibility
| Threshold | |||
|---|---|---|---|
| 8% | 2.5% | 0.8% | |
| Sensitivity | 69% | 86% | 91% |
| Specificity | 100% | 100% | 91% |
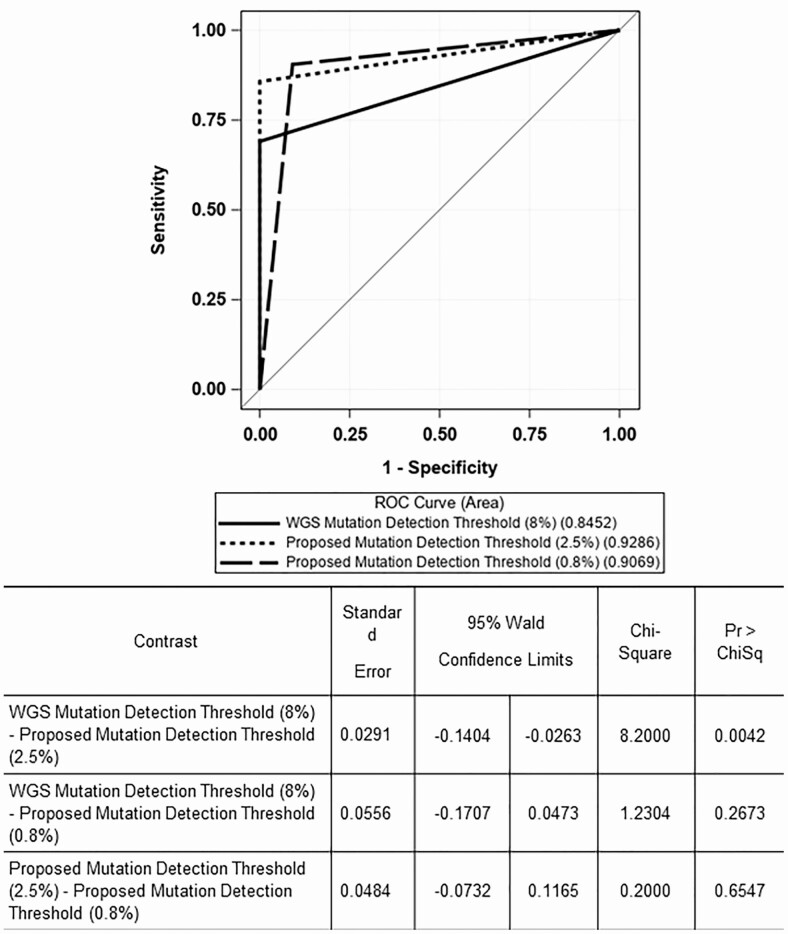
We performed a third ROC curve analysis (Figure 3) to determine which threshold had the highest AUC. We compared the predominant mutation allele frequency threshold observed in the genome-wide analysis (8%) with the 2 predominant mutation allele frequency thresholds determined by the threshold optimality analysis (2.5% and 0.8%). The predominant mutation allele frequency of 2.5% had the highest AUC at 0.93, significantly higher than the 8% threshold (AUC = 0.85; P = .042). The differences in AUC between the 8% and 0.8% predominant mutation allele frequencies and between the 2.5% and 0.8% were not significantly different. These results were confirmed by the McNemar’s test of sensitivities used to compare 2 diagnostic tests. The sensitivities of the diagnostic tests using the 8% and 2.5% predominant allele frequency thresholds were significantly different (P = .016) (Table 7). The McNemar’s test of sensitivities also showed a statistically significant difference between the sensitivities of the 8% and 0.8% thresholds (P = .004) (Table 7), but not between the 2.5% and 0.8% thresholds (P = .5).
Figure 3.
Comparison of mutation allele frequency thresholds for fluoroquinolone resistance. Abbreviations: ROC, receiver operating characteristic; WGS, whole-genome sequencing.
Table 7.
Sensitivity and Specificity of Whole-Genome Sequencing (WGS; Predominant Mutation Allele Frequency) Compared to Phenotypic Drug Susceptibility Testing (DST; Reference Standard)
| Comparison of Sensitivities of the 8% and 2.5% Thresholds | |||||
|---|---|---|---|---|---|
| WGS at 2.5% Threshold | WGS at 8% Threshold | ||||
| Resistant | Not Resistant | Total | P a | ||
| Resistant | 29 | 7 | 36 | .016 | |
| Not Resistant | 0 | 6 | 6 | ||
| Total | 29 | 13 | 42 | ||
| Comparison of Sensitivities of the 8% and 0.8% Thresholds | |||||
| WGS at 0.8% Threshold | WGS at 8% Threshold | ||||
| Resistant | Not Resistant | Total | P a | ||
| Resistant | 29 | 9 | 38 | .004 | |
| Not Resistant | 0 | 4 | 4 | ||
| Total | 29 | 13 | 42 | ||
| Comparison of Sensitivities of the 2.5% and 0.8% Thresholds | |||||
| WGS at 0.8% Threshold | WGS at 2.5% Threshold | ||||
| Resistant | Not Resistant | Total | P a | ||
| Resistant | 36 | 2 | 38 | .5 | |
| Not Resistant | 0 | 4 | 4 | ||
| Total | 36 | 6 | 42 | ||
Abbreviations: DST, drug susceptibility testing; WGS, whole genome sequencing.
a P value derived from McNemar’s test.
DISCUSSION
There were several important findings of this study. First, the proportion of fluoroquinolone-resistant isolates without DNA gyrase mutations identified on whole-genome sequencing (31%) was high and consistent with previous reports [2]. These findings support recent studies demonstrating the need for more sensitive genotypic resistance tests to detect fluoroquinolone resistance in M. tuberculosis [10, 11, 37–39]. Second, widely available pipelines miss phenotypic fluoroquinolone resistance in isolates that have low-level allele frequencies (microheteroresistance); therefore, allele frequency analysis after undergoing rigorous quality-control procedures could be used in conjunction with whole-genome sequencing to improve sensitivity. Third, the predominant DNA gyrase mutation allele frequency provided greater discriminating capacity than the mutational burden in gyrA, gyrB, or the combination of gyrA plus gyrB. Fourth, an allele frequency mutation detection threshold of 2.5% provided significantly greater sensitivity for detecting resistance than whole-genome sequencing analysis done with a widely available pipeline; sensitivity increased from 69% to 86% without sacrificing specificity (Table 6). This approach provides a more sensitive way of detecting fluoroquinolone resistance, especially on heteroresistant strains that are missed by the detection limits of many phenotypic and molecular assays [40]. It should be noted that this occurred with an average depth of coverage of 868 reads.
Importantly, even with more sensitive diagnostic tests, in this cohort of 42 ofloxacin-resistant M. tuberculosis isolates, 6 (14%) would still not be detected by genotypic methods using the 2.5% threshold and 4 (10%) isolates would not have been detected by using 0.8%. This suggests that mechanisms of fluoroquinolone resistance in M. tuberculosis other than those mediated by DNA gyrase mutations may be involved. If alternative resistance mechanisms and associated mutations are identified, they could potentially be included in diagnostic tests to further increase sensitivity.
The predominance of gyrA D94G, D94N, A90V, and S91P mutations in ofloxacin-resistant isolates is consistent with previously published studies [5].
There were several limitations of this study. First, this relatively small sample of ofloxacin-resistant isolates in Tennessee was of predominantly European-American lineage, with few East-Asian strains. The study may have been underpowered to detect novel resistance-conferring mutations since genetic background has been reported to affect both ofloxacin MIC and ofloxacin resistance mutational profiles [41] and could have affected the optimal predominant mutation allele frequency for detecting fluoroquinolone resistance. It is unclear if these results are generalizable to other settings. Second, the list of mutations associated with fluoroquinolone resistance, and the selected genomic positions used in our allele-level analysis, may not include all fluoroquinolone-resistance–conferring mutations. This list was generated by reviewing the current literature and available databases. The list of specific loci, and their association with fluoroquinolone resistance in M. tuberculosis, is constantly evolving. Third, there can be variability in MIC determination. However, each fluoroquinolone-resistant isolate was tested in triplicate by REMA, and we used the standard ofloxacin cutoff of 2 mg/L to determine fluoroquinolone resistance.
A strength of this study was that clinical M. tuberculosis isolates were studied, and thus of greater clinical relevance than laboratory strains. The study also evaluated the entire genome and utilized lineage-matched ofloxacin-susceptible control isolates to increase the likelihood of identifying SNPs associated with fluoroquinolone resistance.
In summary, current whole-genome sequencing analyses are insufficiently sensitive to detect fluoroquinolone resistance in M. tuberculosis, including in heteroresistant clinical isolates. Our study showed that use of the predominant mutation allele frequency in DNA gyrase, in conjunction with whole-genome sequencing, maintains high specificity and also improves the sensitivity to detect fluoroquinolone resistance in M. tuberculosis. Future studies to evaluate the 2.5% threshold as the optimal threshold to detect fluoroquinolone resistance in a more diverse sample are warranted.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the Michigan Department of Community Health for spoligotyping and mycobacterial interspersed repetitive-unit, the Tennessee Public Health Laboratory for the clinical Mycobacterium tuberculosis isolates, and the Tennessee Department of Health for their support of this collaboration. The authors also thank David Sherman, Tige Rustad, and Robert Morrison at the Center for Infectious Disease Research, Seattle, Washington, for their scientific review and advice.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant numbers R56 AI118361, R01 AI063200, K24 065298, K01 AI131895, K08 AI106420).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Piton J, Petrella S, Delarue M, et al. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS One 2010; 5:e12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maruri F, Sterling TR, Kaiga AW, et al. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother 2012; 67:819–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camus JC, Pryor MJ, Médigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 2002; 148:2967–73. [DOI] [PubMed] [Google Scholar]
- 4. Pantel A, Petrella S, Veziris N, et al. Extending the definition of the GyrB quinolone resistance-determining region in Mycobacterium tuberculosis DNA gyrase for assessing fluoroquinolone resistance in M. tuberculosis. Antimicrob Agents Chemother 2012; 56:1990–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avalos E, Catanzaro D, Catanzaro A, et al. Frequency and geographic distribution of gyrA and gyrB mutations associated with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolates: a systematic review. PLoS One 2015; 10:e0120470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coll F, McNerney R, Preston MD, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med 2015; 7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zimenkov DV, Antonova OV, Kuz’min AV, et al. Detection of second-line drug resistance in Mycobacterium tuberculosis using oligonucleotide microarrays. BMC Infect Dis 2013; 13:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McNerney R, Clark TG, Campino S, et al. Removing the bottleneck in whole genome sequencing of Mycobacterium tuberculosis for rapid drug resistance analysis: a call to action. Int J Infect Dis 2017; 56:130–5. [DOI] [PubMed] [Google Scholar]
- 9. Eilertson B, Maruri F, Blackman A, Herrera M, Samuels DC, Sterling TR. High proportion of heteroresistance in gyrA and gyrB in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 2014; 58:3270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campbell PJ, Morlock GP, Sikes RD, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2011; 55:2032–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodwell TC, Valafar F, Douglas J, et al. Predicting extensively drug-resistant Mycobacterium tuberculosis phenotypes with genetic mutations. J Clin Microbiol 2014; 52:781–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Metcalfe JZ, Streicher E, Theron G, et al. Cryptic microheteroresistance explains mycobacterium tuberculosis phenotypic resistance. Am J Respir Crit Care Med 2017; 196:1191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mazars E, Lesjean S, Banuls AL, et al. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci USA 2001; 98:1901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin A, Camacho M, Portaels F, Palomino JC. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob Agents Chemother 2003; 47:3616–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin A, Paasch F, Docx S, et al. Multicentre laboratory validation of the colorimetric redox indicator (CRI) assay for the rapid detection of extensively drug-resistant (XDR) Mycobacterium tuberculosis. J Antimicrob Chemother 2011; 66:827–33. [DOI] [PubMed] [Google Scholar]
- 16. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo Y, Li CI, Sheng Q, et al. Very low-level heteroplasmy mtDNA variations are inherited in humans. J Genet Genomics 2013; 40:607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Starks AM, Avilés E, Cirillo DM, et al. Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform. Clin Infect Dis 2015; 61(Suppl 3):S141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malik S, Willby M, Sikes D, Tsodikov OV, Posey JE. New insights into fluoroquinolone resistance in Mycobacterium tuberculosis: functional genetic analysis of gyrA and gyrB mutations. PLoS One 2012; 7:e39754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ali A, Hasan R, Jabeen K, Jabeen N, Qadeer E, Hasan Z. Characterization of mutations conferring extensive drug resistance to Mycobacterium tuberculosis isolates in Pakistan. Antimicrob Agents Chemother 2011; 55:5654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaoui I, Oudghiri A, El Mzibri M. Characterization of gyrA and gyrB mutations associated with fluoroquinolone resistance in Mycobacterium tuberculosis isolates from Morocco. J Glob Antimicrob Resist 2018; 12:171–4. [DOI] [PubMed] [Google Scholar]
- 25. Chen J, Chen Z, Li Y, et al. Characterization of gyrA and gyrB mutations and fluoroquinolone resistance in Mycobacterium tuberculosis clinical isolates from Hubei Province, China. Braz J Infect Dis 2012; 16:136–41. [DOI] [PubMed] [Google Scholar]
- 26. Chien JY, Chiu WY, Chien ST, Chiang CJ, Yu CJ, Hsueh PR. Mutations in gyrA and gyrB among fluoroquinolone- and multidrug-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 2016; 60:2090–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chernyaeva E, Fedorova E, Zhemkova G, Korneev Y, Kozlov A. Characterization of multiple and extensively drug resistant Mycobacterium tuberculosis isolates with different ofloxacin-resistance levels. Tuberculosis (Edinb) 2013; 93:291–5. [DOI] [PubMed] [Google Scholar]
- 28. Farhat MR, Jacobson KR, Franke MF, et al. Gyrase mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J Clin Microbiol 2016; 54:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McGrath M, Gey van Pittius NC, Sirgel FA, Van Helden PD, Warren RM. Moxifloxacin retains antimycobacterial activity in the presence of gyrA mutations. Antimicrob Agents Chemother 2014; 58:2912–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Streicher EM, Bergval I, Dheda K, et al. Mycobacterium tuberculosis population structure determines the outcome of genetics-based second-line drug resistance testing. Antimicrob Agents Chemother 2012; 56:2420–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Z, Xie T, Mu C, et al. Molecular characteristics of ofloxacin mono-resistant Mycobacterium tuberculosis isolates from new and previously treated tuberculosis patients. J Clin Lab Anal 2018; 32:e22202. doi: 10.1002/jcla.22202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu C, Zhang Y, Shen Y, et al. Molecular characterization of fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates from Shanghai, China. Diagn Microbiol Infect Dis 2012; 73:260–3. [DOI] [PubMed] [Google Scholar]
- 33. Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med 2009; 6:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 1983; 148:839–43. [DOI] [PubMed] [Google Scholar]
- 35. Kumar R, Indrayan A. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr 2011; 48:277–87. [DOI] [PubMed] [Google Scholar]
- 36. Habibzadeh F, Habibzadeh P, Yadollahie M. On determining the most appropriate test cut-off value: the case of tests with continuous results. Biochem Med (Zagreb) 2016; 26:297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miotto P, Cirillo DM, Migliori GB. Drug resistance in Mycobacterium tuberculosis: molecular mechanisms challenging fluoroquinolones and pyrazinamide effectiveness. Chest 2015; 147:1135–43. [DOI] [PubMed] [Google Scholar]
- 38. Farhat MR, Sultana R, Iartchouk O, et al. Genetic determinants of drug resistance in mycobacterium tuberculosis and their diagnostic value. Am J Respir Crit Care Med 2016; 194:621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seifert M, Capparelli E, Catanzaro DG, Rodwell TC. Using mycobacterium tuberculosis single-nucleotide polymorphisms to predict fluoroquinolone treatment response. Antimicrob Agents Chemother 2019; 63:e00076–19. doi: 10.1128/AAC.00076-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rigouts L, Coeck N, Gumusboga M, et al. Specific gyrA gene mutations predict poor treatment outcome in MDR-TB. J Antimicrob Chemother 2016; 71:314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Castro RAD, Ross A, Kamwela L, et al. The genetic background modulates the evolution of fluoroquinolone-resistance in Mycobacterium tuberculosis. Mol Biol Evol 2020; 37:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.