Figure 3.
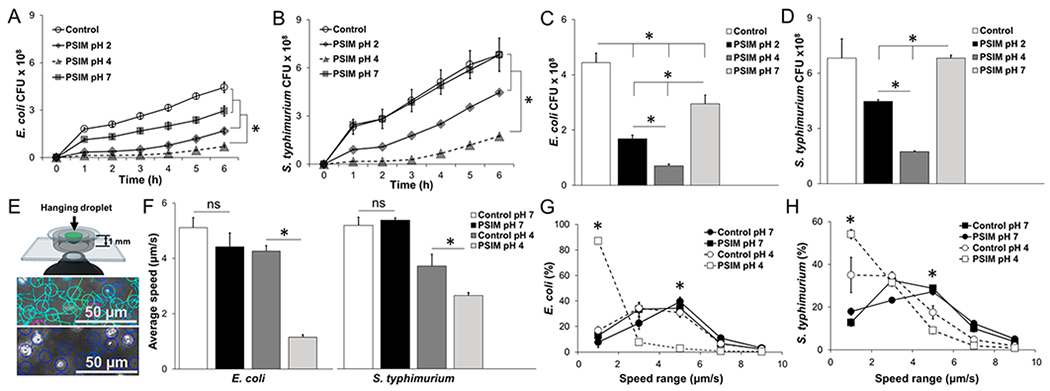
Barrier function of PSIM. (A) Time-resolved transmigration of E. coli through 3 μm pore transwell membranes layered with PSIM at pH 7, 4, and 2. Controls were HEPES buffer at pH 7 without PSIM (n = 3). (B) Transmigration of Salmonella. (C) Number of E. coli that migrated across the PSIM layers at a time point of 6 h. (D) Number of Salmonella that migrated across the PSIM layers at 6 h. (E) The speeds of individual bacteria were measured in hanging drops of PSIM mixed with bacteria and suspended above the objective of an inverted microscope. At pH 7 (top), velocities (indicated by line lengths) were greater than those observed at pH 4 (bottom), where all bacteria were visibly nonmotile. (F) The average speeds of E. coli and Salmonella in PSIM at pH 4 were significantly lower than PSIM-free controls at pH 4 (*, P < 0.05; n = 3). At pH 7, PSIM had no effect on bacterial motility, compared to control. (G) Distribution of speeds of E. coli in PSIM at pH 4 and 7, compared to controls (HEPES buffer without PSIM; n = 3). The percentage of bacteria with speeds of <2 μm/s was greater in PSIM at pH 4, compared to pH 7 and controls (*, P < 0.05). The percentage of bacteria swimming at 5 μm/s was lower in PSIM at pH 4, compared to that observed at pH 7 and for the controls (*, P < 0.05). (H) The distribution of speeds of Salmonella in PSIM at pH 4 and 7, compared to controls was functionally similar to E. coli.
