FIGURE 2.
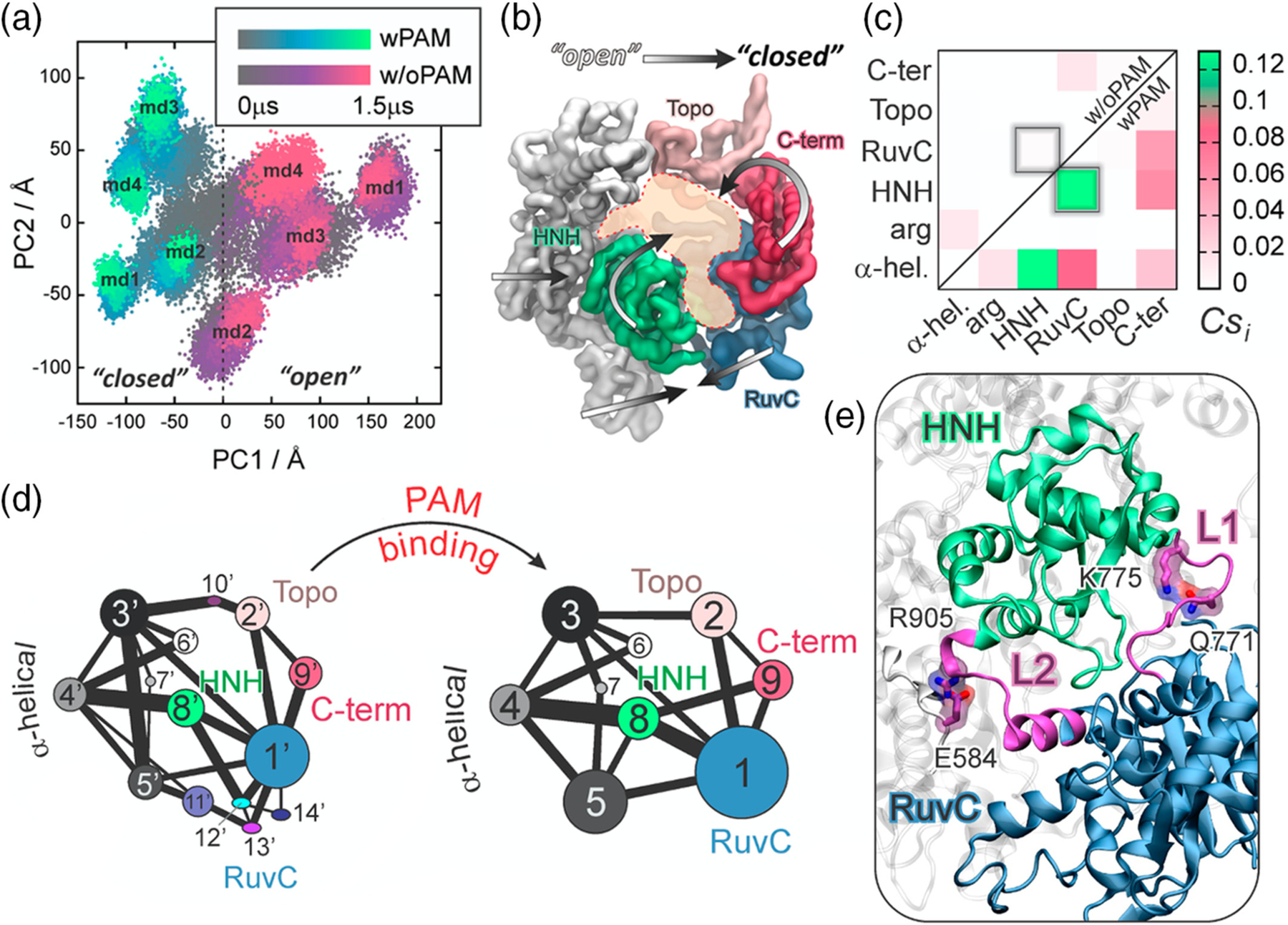
PAM-induced allostery in CRISPR-Cas9. (a) Conformational space adopted by the CRISPR-Cas9 complex spanning over the two principal components of motions (i.e., PC1 vs. PC2), computed over eight independent MD runs of Cas9 bound to PAM (i.e., wPAM) and without PAM (i.e., w/oPAM). (b) “Open-to-close” conformational transition identified along the first principal component. (c) Per-domain correlation score (Csi) matrix, identifying the Cas9 inter-domain coupled motions color-coded green (correlated) to white (not correlated). (d) Community network graph of Cas9–w/oPAM (right) and wPAM (left). Bonds connecting communities correspond to the interconnection strength. (e) Allosteric transducer loops L1/L2 connecting the catalytic HNH and RuvC domain. Critical network nodes (Q771, E584, K775, and R905) of the L1/L2 loops forming essential edges in the allosteric transmission between HNH and RuvC.15 Reprinted with permission from Reference 15 Copyright 2017 American Chemical Society. https://pubs.acs.org/doi/10.1021/jacs.7b05313. Further permissions related to the material excerpted should be directed to the American Chemical Society
