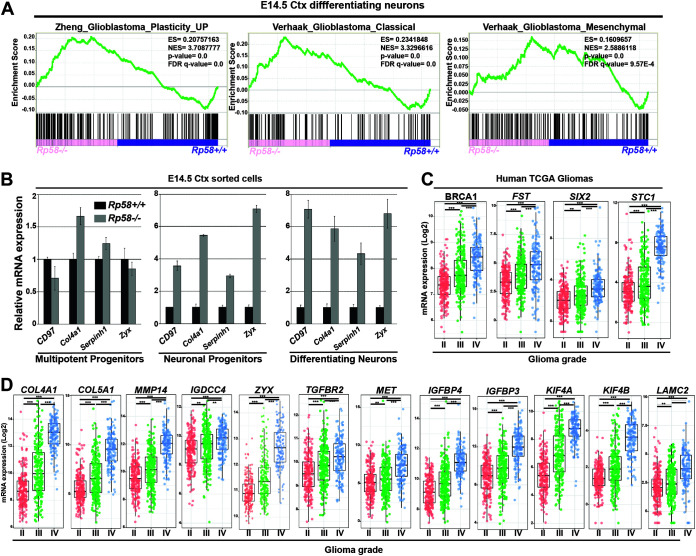
FIG 3.
The genes repressed by RP58 in neuronal cells are associated with gene signatures involved in glioma progression. (A) GSEA analysis shows the significant correlation of the genes upregulated in Rp58−/− neurons with those genes associated with GBM plasticity and GBM classical (C) or mesenchymal (M) subtype. (B) Analyses of the relative mRNA levels of genes involved in the GBM M signature, namely Cd97, Col4a1, Serpinh1, and Zyxin (Zyx), in sorted cell populations of Rp58−/− versus Rp58+/+ E14.5 cortices. Note that the expression of these genes is maintained at a high level in the Rp58−/− cells. Diagrams are from a representative experiment. (C) Box plot showing the normalized expression of BRCA1, FST, SIX2 and STC1 genes in human glioma samples according to tumor grade following analysis of the RNA expression results from The Cancer Genome Atlas (TCGA) GBM–low-grade glioma (LGG) samples using GlioVis (n = 226 grade II, n = 244 grade III, and n = 150 grade IV or GBMs). (D) Box plot showing the normalized expression of RP58 developmental target genes in glioma samples according to tumor grade following analysis of the RNA expression results from TCGA samples using the GlioVis platform (n = 226 grade II, n = 244 grade III, and n = 150 grade IV or GBMs). Gene names are indicated on top of the corresponding box plot. ***, P < 0.001; **, P < 0.01.