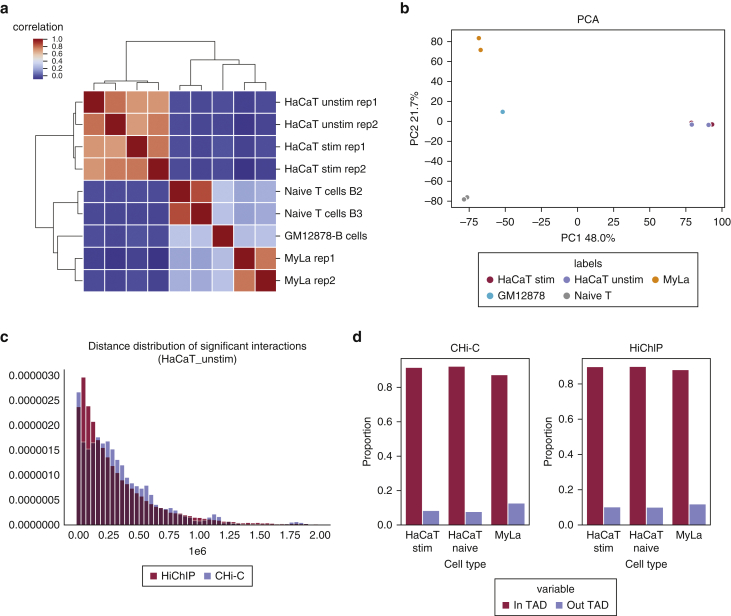
Figure 1.
Properties of the chromatin looping datasets used in this study. (a) Hierarchical clustering of the loops for the individual HiChIP samples using correlation. (b) PCA of the loops for the individual HiChIP samples. (c) Comparison of the distance distribution of the significant fithichip (HiChIP) and CHiCAGO (CHi-C) interactions for naïve HaCaT HiChIP, a representative example. (d) The proportion of significant interactions that are within TADs and that cross TAD boundaries for region capture Hi-C data and HiChIP data. CHi-C, capture Hi-C; PC, principal component; PCA, principal component analysis; rep, replicate; stim, stimulated; TAD, topologically associating domain; unstim, unstimulated.