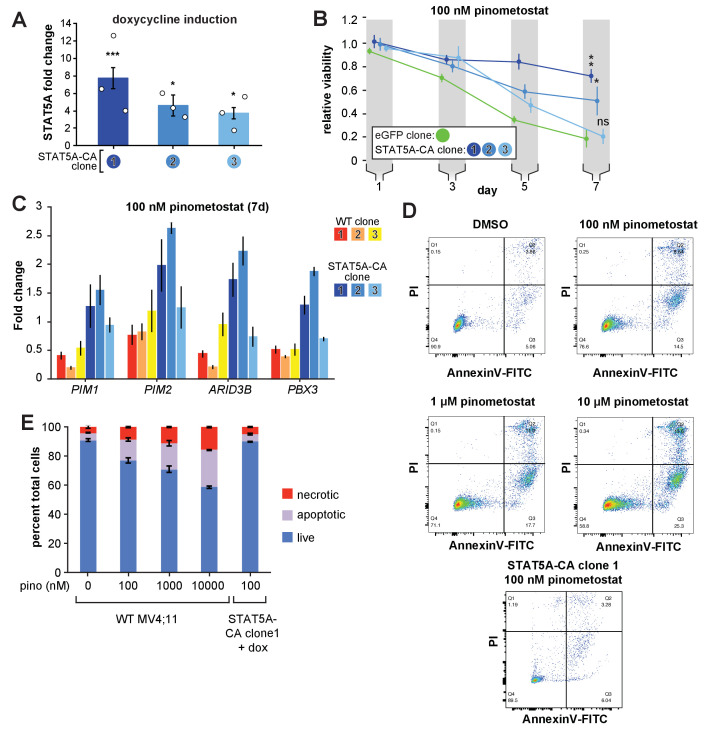
Figure 5. Exogenous expression of constitutively active STAT5A partially rescues proliferation and gene expression effects of DOT1L inhibition.
(A) RT-qPCR analysis of STAT5A expression from three monoclonal isolates of MV4;11 cells virally transduced with a tet-inducible constitutively active STAT5A (STAT5A-CA) depicted as fold-change over untransduced cells with standard error of the mean. Student’s t-test (* p < 0.05, *** p < 0.001). (B) Proliferation assay of MV4;11 clonal isolates from panel A. induced to express STAT5A-CA or eGFP with 1 µg/mL doxycycline and treated concomitantly with 100 nM pinometostat. We determined the fractional viability of each clone as the luminescence from a CellTiter Glo 2.0 assay with pinometostat-treatment normalized to DMSO-treated cells, both induced to express STAT5A-CA or eGFP, to accommodate for any potential increases in viability. Means ± SE are shown for three independent experiments with Student’s t-test for day 7 values (**** p ≤ 0.0001). (C) Gene expression analysis by RT-qPCR of STAT5A target genes in WT MV4;11 cells or MV4;11 STAT5A-CA clones from A. induced with 1 µg/mL doxycycline and treated with 100 nM pinometostat for 7 days. Results are displayed as fold-change over DMSO-treated WT cells. (D) Quantitative measurement by flow cytometry of live, apoptotic (Annexin V-FITC) and necrotic cells (propidium iodide) of WT MV4;11 cells or cells exogenously expressing STAT5A-CA (clone 1) and treated with increasing concentrations of pinometostat. Images of gated FITC vs. PI signal are shown for one of three independent experiments, with all replicates quantified in the bar plot in E.