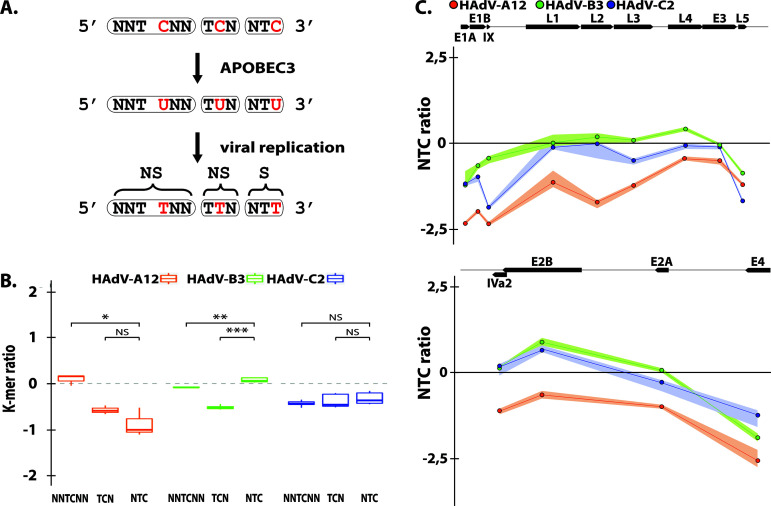
FIG 7.
HAdV-A12 genome bears a strong A3 evolutionary footprint. (A) A3s induce C to U deamination preferentially in a 5′-TC dinucleotide context. After viral replication the mutation is conserved in the viral genome as a C to T transition (in red). Depending on the position of the TC motif in codon, the mutation is synonymous (S) or nonsynonymous (NS). Since synonymous mutations are more likely to be conserved, the A3 footprint is defined as the depletion of the NTC codons. (B) The observed/expected ratios of TC dinucleotide at various codon positions (i.e., NNTCNN, TCN, NTC) were calculated for the HAdV-A12, -B3, and -C2 full-length genomes. Median and quartile are depicted by a boxplot. P values were calculated by Student’s unpaired, two-tailed t test (NS, not significant, *, P < 0.05, **, P < 0.01, ***, P < 0.001. (C) NTC observed/expected ratios were calculated for the different genes of HAdV-A12, -B3, and -C2. Genes’ location, orientation, and length are represented by black arrows.