ABSTRACT
LamPORE is a novel diagnostic platform for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA combining loop-mediated isothermal amplification with nanopore sequencing, which could potentially be used to analyze thousands of samples per day on a single instrument. We evaluated the performance of LamPORE against reverse transcriptase PCR (RT-PCR) using RNA extracted from spiked respiratory samples and stored nose and throat swabs collected at two UK hospitals. The limit of detection of LamPORE was 10 genome copies/μl of extracted RNA, which is above the limit achievable by RT-PCR, but was not associated with a significant reduction of sensitivity in clinical samples. Positive clinical specimens came mostly from patients with acute symptomatic infection, and among them, LamPORE had a diagnostic sensitivity of 99.1% (226/228; 95% confidence interval [CI], 96.9% to 99.9%). Among negative clinical specimens, including 153 with other respiratory pathogens detected, LamPORE had a diagnostic specificity of 99.6% (278/279; 98.0% to 100.0%). Overall, 1.4% (7/514; 0.5% to 2.9%) of samples produced an indeterminate result on first testing, and repeat LamPORE testing on the same RNA extract had a reproducibility of 96.8% (478/494; 94.8% to 98.1%). LamPORE has a similar performance as RT-PCR for the diagnosis of SARS-CoV-2 infection in symptomatic patients and offers a promising approach to high-throughput testing.
KEYWORDS: diagnosis, LamPORE, nanopore sequencing, SARS-CoV-2
INTRODUCTION
Rapid, reliable, high-throughput methods of testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection would help to control transmission. Present diagnosis relies mostly on reverse transcriptase PCR (RT-PCR), but it has proven difficult to expand to the scale needed for population-wide testing of symptomatic individuals. For example, shortages of laboratory RT-PCR capacity still limit the United Kingdom testing program over a year after the SARS-CoV-2 pandemic was declared a public health emergency of international concern by the WHO. Further expansion of testing to include screening of asymptomatic individuals, which may be needed to prevent SARS-CoV-2 circulation, would require a significant further increase in testing capacity (1, 2).
In the United Kingdom, clinical laboratories have struggled to expand conventional RT-PCR workflows to meet the demand for SARS-CoV-2 testing, and many have explored alternative methods that would be more scalable or allow near-patient use (3, 4). At the Oxford University Hospitals NHS Foundation Trust (OUH) and Sheffield Teaching Hospitals NHS Foundation Trust (STH), we evaluated LamPORE, a novel diagnostic platform for SARS-CoV-2 developed by Oxford Nanopore Technologies (ONT) that combines loop-mediated isothermal amplification (LAMP) with nanopore sequencing (5). During sample preparation, a unique combination of DNA barcodes are incorporated into the LAMP products from each specimen so that they can be pooled into a single sequencing run. In the current protocol, up to 92 samples can be analyzed on 1 flow cell, potentially allowing thousands of samples to be analyzed per day on a single instrument running multiple flow cells in parallel. The workflow involves a 40-minute amplification, followed by library preparation, and a 60-minute sequencing run, generating results in a comparable time to RT-PCR when starting with extracted RNA.
As well as molecular barcoding, using sequencing to detect the outcome of the LAMP reaction offers other advantages compared with simpler LAMP assays that detect the presence of DNA synthesis by measurement of pH, turbidity, or fluorescent dyes. Sequenced reads from a specific target will contain sequences not present in the primers, avoiding false positives caused by nonspecific amplification (although the amplicons are not large enough to usefully genotype the virus) (6). Conversely, reads confidently assigned to SARS-CoV-2 targets may indicate a true positive even if present at relatively low levels, potentially improving the low sensitivity seen in several LAMP assays compared with RT-PCR (7, 8). In addition, the detection of LAMP products by sequencing allows the possibility of multiplexing the assay with other pathogens. LamPORE uses ONT flow cells compatible with several sequencing instruments, including the portable MinION device and high-throughput GridION and PromethION platforms, and thus, it could potentially be used both for mobile and centralized testing.
In this evaluation, we compare the performance of LamPORE with RT-PCR on extracted RNA from respiratory specimens. Initially, we use spiked samples to determine the analytical limit of detection of the assay. We then use stored clinical samples to determine the assay’s diagnostic sensitivity, specificity, and reproducibility.
MATERIALS AND METHODS
The evaluation was conducted across three sites, namely, OUH, STH, and the Public Health England National Infection Service at Porton Down (PHE Porton Down).
LamPORE.
LamPORE is a CE marked diagnostic assay developed by ONT and described in detail in James, et al. (5). The assay was performed identically at each site using a GridION instrument with operators unaware of reference PCR results. It takes 20-μl RNA input into a single multiplex reaction targeting the following three regions of the SARS-CoV-2 genome using previously published primers (9): ORF1a, envelope, and nucleocapsid genes, plus human β-actin mRNA as a control of sampling adequacy and assay performance. LamPORE sample preparation uses a 96-well plate format, with each sample having 1 of 8 LAMP forward inner primer (FIP) barcodes and 1 of 12 transposase (rapid) barcodes added before pooling. In these experiments, a single LAMP barcode (FIP7) was not used, as it had previously been associated with lower β-actin read counts and was awaiting replacement (unpublished data). As a result, plates contained 80 samples, plus 2 no-template controls and 2 positive controls consisting of synthetic SARS-CoV-2 RNA (Twist Bioscience). To assess for potential sample-sample contamination, positive and negative clinical samples were intermixed, with positions altered between replicates.
We used the LamPORE protocol dated 1 July 2020 (version 1, revision 4). Briefly, this protocol consists of adding sample RNA to LAMP master mix and primers and then incubating the mixture at 65°C to 80°C in a thermocycler for 40 minutes, during which time amplification occurs and the LAMP primer barcodes are incorporated into concatemers containing the target sequence. Following these steps, samples from the same column are pooled and a second set of barcodes are incorporated using a rapid transposase-based method. All samples are then pooled into a single sequencing library, with no need for normalization, as DNA concentrations are similar in all positive samples following LAMP, regardless of the initial viral load. The pooled library has a magnetic bead cleanup, then is added to a MinION flow cell, and sequenced for 60 minutes, after which a report is generated automatically by the instrument within seconds for each barcode set. Unlike RT-PCR, LamPORE does not provide the equivalent of a cycle threshold (CT) value reflecting the initial viral load, as measurement occurs only after amplification is complete. The number of reads assigned to each target is used to generate a report as follows: (i) invalid, <50 classified reads in total detected from SARS-CoV-2 and β-actin targets, (ii) positive, ≥50 SARS-CoV-2 reads detected (adding read counts across all three SARS-CoV-2 targets), (iii) Inconclusive, not invalid and ≥20 and <50 SARS-CoV-2 reads detected, and (iv) negative, not invalid and <20 SARS-CoV-2 reads detected.
Spiked samples—PHE Porton Down.
Spiked samples were prepared and analyzed at PHE Porton Down to establish the limits of detection of LamPORE. Aliquots of pooled volunteer saliva were used for spiking experiments, which were confirmed SARS-CoV-2 negative by RT-PCR. They were spiked with cultured SARS-CoV-2 (Victoria/01/202026 passaged twice in Vero/hSLAM cells) at 1,000 SARS-CoV-2 genome copies/ml of sample and serially diluted with the remaining material to create a dilution series of positive samples.
RNA was extracted from 360 μl of the spiked sample using the QiaAMP viral RNA minikit (Qiagen), with RNA eluted in 36 μl. Reference RT-PCR was conducted with the CDC NS1 assay with 5-μl RNA input (10). Quantification was determined by comparison to a standard curve of a plasmid 2019-nCoV_N positive control (Integrated DNA Technologies). Further details are in the supplemental material.
Clinical specimens—OUH and STH.
Testing of stored clinical samples was performed at OUH and STH. All samples were nose and/or throat swabs collected into viral transport media during routine clinical care and stored at −80°C.
(i) Sample selection.
(a) SARS-CoV-2-positive samples. At OUH, sequentially available positive specimens collected from March to April 2020 were chosen without reference to the RT-PCR cycle threshold (CT) value. During this time, a uniplex RdRp RT-PCR assay was in use, based on the assay described by Corman et al. (11). At STH, a stratified random sample of specimens collected from April to May 2020 were selected based on their initial SARS-CoV-2 E gene CT value, using an in-house assay based on the Corman et al. protocol (11, 12), with 50% chosen to have CT values of <30 and 50% to have ≥30. At both sites, testing was largely restricted to hospitalized patients and symptomatic staff during the collection period.
(b) SARS-CoV-2-negative samples. At OUH, negative samples were selected from stored prepandemic respiratory samples. They had initially been tested with either GeneXpert Flu/RSV (Cepheid) or the BioFire FilmArray respiratory panel 2.0 (bioMérieux) and were purposefully chosen to include samples with a range of other respiratory pathogens. Over 90% of samples were collected between October and December 2019, but those samples containing non-SARS-CoV-2 seasonal coronaviruses were used up until a collection date of 10 March 2020 to increase the number available. At STH, negative samples were selected from among those submitted for SARS-CoV-2 testing.
(ii) RNA extraction.
For samples originating from OUH, RNA extraction was conducted with the QIAsymphony SP instrument and the DSP virus/pathogen kit (Qiagen) (13). A total of 200 μl of viral transport medium was extracted, and RNA was eluted in 60 μl. For samples originating from STH, RNA extraction was performed using the MagNA Pure 96 instrument with the MagNA Pure 96 DNA and viral neuraminidase (NA) small volume kit (Roche). A total of 200 μl of viral transport medium was extracted, and RNA was eluted in 100 μl. Aliquots of RNA were stored at −80°C prior to analysis.
(iii) Reference RT-PCR.
Reference RT-PCR was undertaken contemporaneously with LamPORE on aliquots of the same RNA extract, with operators unaware of LamPORE results. For samples originating from OUH, the reference RT-PCR was the RealStar SARS-CoV-2 RT-PCR assay (Altona Diagnostics) using 10-μl RNA input. For samples originating from STH, an in-house RT-PCR assay based on Corman et al. methods was used with 6-μl RNA input (11, 12). Further details are in the supplemental material.
(iv) Replicates.
To assess the reproducibility of the assay, LamPORE replicates were performed on aliquots of the same RNA extract. To ensure comparable RT-PCR and LamPORE results between OUH and STH, a subset of samples was exchanged between sites, with LamPORE and reference RT-PCR repeated.
Statistical analysis.
R version 3.5.0 was used for analysis with exact binomial confidence intervals calculated for proportions. Initial LamPORE replicates were used to derive estimates of sensitivity and specificity, with second replicates used to estimate LamPORE reproducibility. Results are reported in line with the Standards for Reporting Diagnostic accuracy studies (a STARD checklist is in the supplemental material).
Ethics.
The process for collection of the donated saliva was approved by the PHE Research Ethics and Governance Group. The protocol for the use of stored clinical samples at OUH and STH was reviewed by the Institutional Review Board of OUH and the University of Oxford, and it was determined that the activity constituted service evaluation and service development. As such, it did not need research ethics review.
RESULTS
Limit of Detection.
Using samples spiked with cultured SARS-CoV-2, LamPORE had a limit of detection of 1,000 SARS-CoV-2 genome copies/ml of sample and detected 15/15 samples (Table 1). With the RNA extraction protocol used, and assuming 100% extraction efficiency, this limit of detection would correspond to a concentration of 10 genome copies/μl of extracted RNA (or 200 copies per 20-μl reaction). Although LamPORE did not consistently detect spiked samples at concentrations below this value, it was positive in 8/18 (44%) samples at a concentration of 100 copies/ml of sample, corresponding to 1 genome copy/μl of extracted RNA (or 20 copies per 20-μl reaction). By comparison, RT-PCR using the CDC NS1 assay was also positive in 15/15 samples at 1,000 SARS-CoV-2 genome copies/ml of sample and in 14/18 (78%) of samples at 100 copies/ml of sample, although the difference with LamPORE was not statistically significant (P = 0.09 by Fisher’s exact test).
TABLE 1.
Limit of detection of LamPORE using spiked samplesa
| SARS-CoV-2 genome copiesb |
LamPORE results (n) |
RT-PCR results |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of replicates | Per ml sample | Per 20-μl LamPORE reaction | Per 5-μl RT-PCR reaction | Per μl of extracted RNA | Positive | Negative | Inconclusive/invalid | RT-PCR positive (n) | Mean CT value ± SD |
| 15 | 1,000 | 200 | 50 | 10 | 15 | 0 | 0 | 15/15 | 32.1 ± 0.5 |
| 18 | 100 | 20 | 5 | 1 | 8 | 10 | 0 | 14/18 | 35.7 ± 1.0 |
| 18 | 10 | 2 | 0.5 | 0.1 | 0 | 18 | 0 | 0/18 | NDc |
| 15 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0/15 | ND |
Copies/RT-PCR is calculated for the comparator CDC NS1 RT-PCR assay using 5-μl RNA input volume.
The relationship between copies/ml sample and copies/μl extracted RNA applies to the extraction method used here, in which RNA from a 360-μl sample was eluted in 36 μl.
ND, not determined.
Diagnostic performance.
Diagnostic performance of LamPORE was assessed using 514 stored nose and throat swabs, 400 from OUH and 114 from STH (details in Table S1 in the supplemental material). Requesting location was available for 135/150 (90%) SARS-CoV-2-positive samples from OUH but not for other samples. Among these samples, 41 (30%) were from outpatient locations (including occupational health), 24 (18%) were from community hospitals, 45 (33%) were from emergency departments or acute admission wards, and 25 (19%) were from other inpatient locations. Sixty cross-site replicates demonstrated good correlation between RT-PCR CT values for E gene targets at OUH and STH despite different assays being used, so this was used as the reference CT (see Fig. S1 in the supplemental material). Samples were analyzed on a total of 13 LamPORE runs performed on separate days.
Among 229 RT-PCR-positive samples tested by LamPORE, 226 were reported positive and 2 were reported negative, giving an overall diagnostic sensitivity of 99.1% (226/228; 95% CI, 96.9% to 99.9%) (Table 2). All valid samples at CT values of 34.9 or lower were positive by LamPORE (Table 3). Considering performance at lower viral loads, 7/9 samples with a CT value of ≥35 were positive and 22/22 of those with CT values between 30 and 34.9 were positive. Both false-negative samples by LamPORE had CT values of ≥38, and 1 of them was positive by LamPORE on repeat testing (see Table S2 in the supplemental material). The one RT-PCR-positive sample that was invalid on initial LamPORE testing was correctly positive when repeated.
TABLE 2.
Clinical diagnostic performance of LamPORE vs RT-PCR
| RT-PCR result | LamPORE result (n) |
||||
|---|---|---|---|---|---|
| Positive | Negative | Inconclusive | Invalid | Total | |
| Positive | 226 | 2 | 0 | 1 | 229 |
| Negative | 1 | 278 | 3 | 3 | 285 |
| Total | 227 | 280 | 3 | 4 | 514 |
TABLE 3.
Performance of LamPORE in the SARS-CoV-2-positive clinical samples by RT-PCR E gene CT value
| RT-PCR CT value | LamPORE result (n) |
||||
|---|---|---|---|---|---|
| Total | Positive | Negative | Inconclusive | Invalid | |
| <15 | 23 | 23 | 0 | 0 | 0 |
| 15.0–19.9 | 51 | 51 | 0 | 0 | 0 |
| 20.0–24.9 | 73 | 72 | 0 | 0 | 1 |
| 25.0–29.9 | 51 | 51 | 0 | 0 | 0 |
| 30.0–34.9 | 22 | 22 | 0 | 0 | 0 |
| >35 | 9 | 7 | 2 | 0 | 0 |
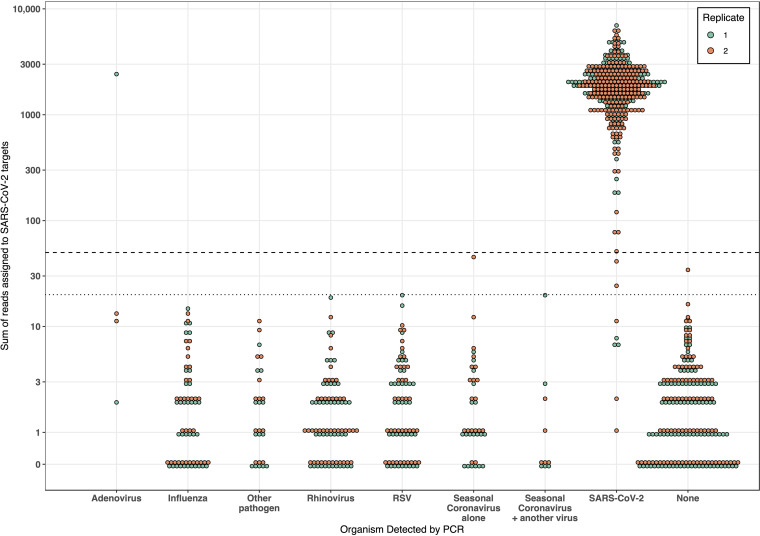
Of 285 RT-PCR-negative samples, 278 were negative and 1 was positive by LamPORE, giving an overall diagnostic specificity of 99.6% (278/279; 98.0% to 100.0%) (Table 2). The false positive was a prepandemic respiratory sample that was also positive for adenovirus and which had 2,419 SARS-CoV-2 reads detected. However, this sample was negative on repeat LamPORE testing (Fig. 1). Six RT-PCR-negative samples gave indeterminate results (three invalid, three inconclusive), of which four were correctly negative on repeat testing, one remained invalid, and one was not retested. Overall, among both RT-PCR-positive and -negative samples, 1.4% (7/514; 0.5% to 2.9%) produced an indeterminate result on first testing.
FIG 1.
Analytical specificity of LamPORE in samples positive for a range of respiratory pathogens. Data for both LamPORE replicates are shown. The dashed line is the threshold for a positive result (≥50 reads), and the dotted line is the threshold for an inconclusive result (≥20 reads). “Other pathogens” includes parainfluenza virus (n = 10), Mycoplasma pneumoniae (n = 3), and human metapneumovirus (n = 1). Invalid samples are plotted.
Another respiratory pathogen was detected by multiplex RT-PCR in 153 negative samples, including 43 with rhinovirus, 38 with respiratory syncytial virus (RSV), 33 with influenza, and 24 with seasonal coronaviruses (9 HKU1, 7 NL63, 7 OC43, and 1 229E). Overall, there was no evidence that the presence of any other respiratory pathogen was associated with false-positive results or greater numbers of reads assigned to SARS-CoV-2 targets (Fig. 1).
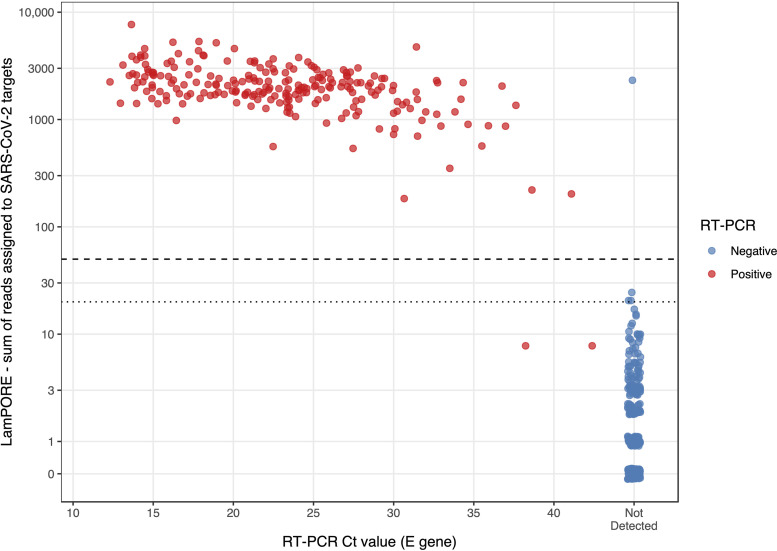
As well as the categorical result produced by the LamPORE reporting algorithm, RT-PCR results were compared with the number of reads assigned by LamPORE to SARS-CoV-2 targets (Fig. 2). This comparison showed that the prespecified cutoff of ≥50 for a positive result was optimal, with any cutoff in the range of 25 to 182 producing a maximal Youden index (sensitivity + specificity − 1) of 0.988. As the rate at which reads are detected becomes roughly constant after a few minutes of sequencing, the effect of a sequencing run longer or shorter than 60 min can be inferred. All samples reported positive by LamPORE had >180 SARS-CoV-2 reads detected and so would have been positive after 30 min of sequencing, at which point there would have been no increase in indeterminate results. Conversely, extending the sequencing duration with the same diagnostic thresholds would not have allowed the detection of either of the two false-negative samples without producing large numbers of false positives.
FIG 2.
Total number of reads assigned to SARS-CoV-2 targets by LamPORE versus RT-PCR E gene CT value. The dashed line is the threshold for a positive result (≥50 reads), and the dotted line is the threshold for an inconclusive result (≥20 reads). Results are for replicate 1 only. Invalid samples are not shown.
Reproducibility.
LamPORE was repeated on 494 samples using the same RNA extract and produced identical results in 478, giving an overall reproducibility of 96.8% (478/494; 94.8% to 98.1%) (Table S3 and S4 in the supplemental material). In four samples (0.8%) with discrepant LamPORE results, the same sample switched between negative and positive. In the other 12 discrepant samples, LamPORE replicates included 1 indeterminate result. All 90 cross-site LamPORE replicates performed between Oxford and Sheffield were concordant (60 RT-PCR/LamPORE positive and 30 RT-PCR/LamPORE-negative). Combining results from replicates to assess the rate of possible sample-sample contamination, 1/576 (0.2%; 0.0% to 1.0%) negative samples or no-template controls were positive by LamPORE.
DISCUSSION
LamPORE has been identified by the UK government as a possible high-throughput platform that could alleviate shortages in SARS-CoV-2 testing capacity (14). In a manuscript released by its developers, LamPORE correctly detected SARS-CoV-2 in 79 of 80 clinical specimens (98.8%; 95% CI, 93.2% to 100.0%), although no SARS-CoV-2-negative specimens were available for testing. Instead, the assay was tested on 85 nonrespiratory human RNA extracts, and 4 were incorrectly reported as positive (sensitivity, 95.2%; 88.3% to 98.7%), which the authors attributed to probable sample contamination (5). In this evaluation, we found that LamPORE had a high diagnostic sensitivity (99.1%) and specificity (99.6%) in our clinical sample set. Combined with a high reproducibility (96.8%) both within and across sites, these results support its practical use for high-throughput testing in a low-prevalence population. Although the assay we evaluated targeted SARS-CoV-2 alone, the LamPORE platform could be adapted to detect other pathogens and is amenable to multiplexing in order to target multiple pathogens in the same assay.
The limit of detection of LamPORE, at 10 genome copies/μl of extracted RNA, was somewhat higher than the 2 copies/μl achievable in previous evaluations of high-performance RT-PCR (15), but this value did not correspond to a significant loss of diagnostic sensitivity in the clinical samples. In our spiking experiments, RNA was extracted from 360 μl transport medium and eluted in 36 μl, a 10-fold concentration. This amount is higher than that of most commonly used extraction protocols, for example, those used at OUH and STH produced 3-fold and 2-fold concentrations, respectively. Therefore, the limit of detection, measured in genome copies/ml of sample, using LamPORE with a high-concentration extraction would be similar to PCR as commonly used with a low-concentration extraction. Automated, commercially available extraction methods can produce a 20-fold RNA concentration, which could further improve the limit of detection, although higher degrees of concentration could lead to assay inhibition, so this would need further evaluation.
Although no clinical metadata were available about the individuals whose samples were used in this evaluation, they would have mainly been derived from patients with acute symptomatic infection, often requiring admission to hospital, as testing was mainly limited to this group during the first wave of infection. The distribution of CT values may be higher in a population with more mild or asymptomatic infections and would be markedly higher among those who remain RT-PCR positive weeks after recovering from acute infection (16). Our data suggest that LamPORE is most likely to miss weakly positive samples with CT values above 35 and thus could have had lower diagnostic sensitivity if tested in such groups. However, this may not be a significant practical disadvantage, as although weak positives have some value for contact tracing, they are likely to come from individuals with low infectious potential (17, 18).
Our evaluation has several limitations. It was conducted after the first wave of COVID-19 in the United Kingdom, when there were few incident cases, so we were unable to prospectively collect samples and instead relied on frozen transport media, which could differ from fresh material. Sample collection occurred at a time when there was little genetic variation in SARS-CoV-2, and we did not attempt to assess the possible effect of future sequence variation causing failure in any of the three gene targets. Positives were defined by a positive RT-PCR at the time of initial sample collection and by repeat positive RT-PCR simultaneously with LamPORE, but although RT-PCR is used as a reference test for SARS-CoV-2, there are many reports of its suboptimal sensitivity in clinical infection (19).
This early evaluation of LamPORE compared its performance against RT-PCR using extracted RNA, as this is the standard material used for the detection of SARS-CoV-2. However, the requirement for viral inactivation and RNA extraction and the additional need for LamPORE library preparation could lead to bottlenecks that would mitigate the potential benefit of LamPORE for high-throughput or mobile testing. LAMP reactions are reported to be more robust than RT-PCR to inhibitors present in clinical samples and so may have superior performance with extraction-free protocols (20, 21). The use of such extraction-free protocols could greatly streamline the workflow, but further evaluation is required. We also did not evaluate how the throughput and turnaround time of LamPORE would compare with RT-PCR during routine use in a clinical laboratory or centralized testing center. The benchtop GridION instrument can accommodate five flow cells simultaneously and so could analyze over 3,000 samples in a 12-hour day at two-thirds occupancy, and the PromethION instrument has a theoretical capacity more than 10-fold higher, but using LamPORE to test tens or hundreds of thousands of samples per day would be dependent on a streamlined workflow, including automated sample handling, integration with laboratory information management systems, and careful safeguards to minimize the risk of contamination.
In conclusion, we show that LamPORE on extracted RNA offers a promising method of high-throughput SARS-CoV-2 testing. However, further evaluation in mild or asymptomatic infection is needed, and large-scale use requires the development of streamlined workflows, possibly by including simpler sample preparation to avoid the need for conventional RNA extraction.
ACKNOWLEDGMENTS
We are grateful to all the clinical microbiology and virology staff at OUH and STH who helped to process the specimens used in this evaluation and to Kevin Bewley, PHE Porton Down, for providing the cultured virus.
Materials for the evaluation were supplied by Oxford Nanopore Technologies, but all experiments and analyses were conducted independently by the investigators.
D.W.E. declares lecture fees from Gilead, outside the submitted work. All other authors declare no competing interests.
This work was supported by the National Institute for Health Research (NIHR) Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance at the University of Oxford in partnership with Public Health England (PHE) and the NIHR Oxford Biomedical Research Centre. L.P. is an NIHR clinical lecturer. D.W.E. is a Robertson Foundation Fellow and Oxford NIHR Senior Research Fellow. P.C.M. is funded by the Wellcome Trust (110110/Z/15/Z). T.I.D.S. is a supported by a Wellcome Trust Intermediate Clinical Fellowship (110058/Z/15/Z). This report presents independent research. The views expressed in this publication are those of the authors and not necessarily those of the NHS, NIHR, the Department of Health, or PHE.
The funding source had no role in study design, in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Conceptualization, M.W.C., D.W.C., T.I.D.S., C.M.E., L.P., and S.T.P.; Methodology, D.P.C., D.W.C., T.I.D.S., C.M.E., L.P., S.T.P., and G.R.; Validation, D.P.C., K.J., K.L.O., M.D.P., M.R., G.R., M.D.W., and M.Y.; Formal analysis, D.W.E., L.P., and T.E.A.P.; Investigation, D.P.C., K.J., K.L.O., M.D.P., L.P., S.T.P., M.R., G.R., M.D.W., and M.Y.; Resources, D.P.C., M.W.C., M.A., D.W.C., T.I.D.S., C.M.E., S.H., A.J., P.C.M., S.T.P., N.S., and A.V.; Writing – Original draft, L.P.; Writing – Review & Editing, all authors; Visualization, L.P.; Supervision, M.W.C., D.W.C., T.I.D.S., C.M.E., and S.T.P.; Project administration, D.W.C., T.I.D.S., C.M.E., S.H., L.P., and S.T.P.; Funding acquisition, M.W.C., D.W.C., T.I.D.S., C.M.E., S.H., T.E.A.P., and S.T.P.
Footnotes
Supplemental material is available online only.
Contributor Information
Leon Peto, Email: leon.peto@ndm.ox.ac.uk.
Michael J. Loeffelholz, Cepheid
REFERENCES
- 1.Grassly NC, Pons-Salort M, Parker EPK, White PJ, Ferguson NM, Ainslie K, Baguelin M, Bhatt S, Boonyasiri A, Brazeau N, Cattarino L, Coupland H, Cucunuba Z, Cuomo-Dannenburg G, Dighe A, Donnelly C, van Elsland SL, FitzJohn R, Flaxman S, Fraser K, Gaythorpe K, Green W, Hamlet A, Hinsley W, Imai N, Knock E, Laydon D, Mellan T, Mishra S, Nedjati-Gilani G, Nouvellet P, Okell L, Ragonnet-Cronin M, Thompson HA, Unwin HJT, Vollmer M, Volz E, Walters C, Wang Y, Watson OJ, Whittaker C, Whittles L, Xi X. 2020. Comparison of molecular testing strategies for COVID-19 control: a mathematical modelling study. Lancet Infect Dis 20:1381–1389. 10.1016/S1473-3099(20)30630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peto J, Carpenter J, Smith GD, Duffy S, Houlston R, Hunter DJ, McPherson K, Pearce N, Romer P, Sasieni P, Turnbull C. 2020. Weekly COVID-19 testing with household quarantine and contact tracing is feasible and would probably end the epidemic. R Soc Open Sci 7:200915. 10.1098/rsos.200915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler VL, Armson B, Gonzales JL, Wise EL, Howson ELA, Vincent-Mistiaen Z, Fouch S, Maltby CJ, Grippon S, Munro S, Jones L, Holmes T, Tillyer C, Elwell J, Sowood A, de Peyer O, Dixon S, Hatcher T, Patrick H, Laxman S, Walsh C, Andreou M, Morant N, Clark D, Moore N, Houghton R, Cortes NJ, Kidd SP. 2021. A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J Infect 82:117–125. 10.1016/j.jinf.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Manzano J, Malpartida-Cardenas K, Moser N, Pennisi I, Cavuto M, Miglietta L, Moniri A, Penn R, Satta G, Randell P, Davies F, Bolt F, Barclay W, Holmes A, Georgiou P. 2021. Handheld point-of-care system for rapid detection of SARS-CoV-2 extracted RNA in under 20 min. ACS Cent Sci 7:307–317. 10.1021/acscentsci.0c01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James P, Stoddart D, Harrington ED, Beaulaurier J, Ly L, Reid SW, Turner DJ, Juul S. 2020. LamPORE: rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv 10.1101/2020.08.07.20161737. [DOI] [Google Scholar]
- 6.Schmid-Burgk JL, Schmithausen RM, Li D, Hollstein R, Ben-Shmuel A, Israeli O, Weiss S, Paran N, Wilbring G, Liebing J, Feldman D, Słabicki M, Lippke B, Sib E, Borrajo J, Strecker J, Reinhardt J, Hoffmann P, Cleary B, Hölzel M, Nöthen MM, Exner M, Ludwig KU, Regev A, Zhang F. 2020. LAMP-seq: population-scale COVID-19 diagnostics using combinatorial barcoding. bioRxiv 10.1101/2020.04.06.025635. [DOI]
- 7.Dao Thi VL, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, Freistaedter A, Papagiannidis D, Galmozzi C, Stanifer ML, Boulant S, Klein S, Chlanda P, Khalid D, Barreto Miranda I, Schnitzler P, Kräusslich HG, Knop M, Anders S. 2020. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med 12:eabc7075. 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rödel J, Egerer R, Suleyman A, Sommer-Schmid B, Baier M, Henke A, Edel B, Löffler B. 2020. Use of the variplex SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J Clin Virol 132:104616. 10.1016/j.jcv.2020.104616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Ren G, Buss J, Barry AJ, Patton GC, Tanner NA. 2020. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques 69:178–185. 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
- 10.FDA. 2020. CDC 2019 novel coronavirus (nCoV) real-time RT-PCR diagnostic panel—instructions for use. https://www.fda.gov/media/134922/download. Accessed 10 September 2020.
- 11.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25:2431. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colton H, Ankcorn M, Yavuz M, Tovey L, Cope A, Raza M, Keeley AJ, State A, Poller B, Parker M, de Silva TI, Evans C. 2020. Improved sensitivity using a dual target, E and RdRp assay for the diagnosis of SARS-CoV-2 infection: experience at a large NHS Foundation Trust in the UK. J Infect 82:159–198. 10.1016/j.jinf.2020.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiagen. QIAsymphony SP protocol sheet Complex200_OBL_V4_DSP protocol. https://www.qiagen.com/gb/resources/resourcedetail?id=02c3171f-9867-43eb-b48b-135afe95b29e&lang=en. Accessed 10 September 2020.
- 14.Roll-out of 2 new rapid coronavirus tests ahead of winter. https://www.gov.uk/government/news/roll-out-of-2-new-rapid-coronavirus-tests-ahead-of-winter. Accessed 22 September 2020.
- 15.FIND. 2021. FIND evaluation update: SARS-CoV-2 molecular diagnostics. Foundation for Innovative New Diagnostics, Geneva, Switzerland. https://www.finddx.org/covid-19/sarscov2-eval-molecular/.
- 16.Rao SN, Manissero D, Steele VR, Pareja J. 2020. A Systematic Review of the Clinical Utility of Cycle Threshold Values in the Context of COVID-19. Infect Dis Ther 9:573–586. 10.1007/s40121-020-00324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, Ladhani S, Zambon M, Gopal R. 2020. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance 25:465. 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jefferson T, Spencer EA, Brassey J, Heneghan C. 2020. Viral cultures for COVID-19 infectious potential assessment—a systematic review. Clin Infect Dis 10.1093/cid/ciaa1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woloshin S, Patel N, Kesselheim AS. 2020. False negative tests for SARS-CoV-2 infection—challenges and implications. N Engl J Med 383:e38. 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko H, Kawana T, Fukushima E, Suzutani T. 2007. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods 70:499–501. 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Tani H, Teramura T, Adachi K, Tsuneda S, Kurata S, Nakamura K, Kanagawa T, Noda N. 2007. Technique for quantitative detection of specific DNA sequences using alternately binding quenching probe competitive assay combined with loop-mediated isothermal amplification. Anal Chem 79:5608–5613. 10.1021/ac070041e. [DOI] [PubMed] [Google Scholar]