ABSTRACT
Immune memory represents the most efficient defense against invasion and transmission of infectious pathogens. In contrast to memory T and B cells, the roles of innate immunity in recall responses remain inconclusive. In this study, we identified a novel mouse spleen NK cell subset expressing NKp46 and NKG2A induced by intranasal influenza virus infection. These memory NK cells specifically recognize N-linked glycosylation sites on influenza hemagglutinin (HA) protein. Different from memory-like NK cells reported previously, these NKp46+ NKG2A+ memory NK cells exhibited HA-specific silence of cytotoxicity but increase of gamma interferon (IFN-γ) response against influenza virus-infected cells, which could be reversed by pifithrin-μ, a p53-heat shock protein 70 (HSP70) signaling inhibitor. During recall responses, splenic NKp46+ NKG2A+ NK cells were recruited to infected lung and modulated viral clearance of virus and CD8+ T cell distribution, resulting in improved clinical outcomes. This long-lived NK memory bridges innate and adaptive immune memory response and promotes the homeostasis of local environment during recall response.
IMPORTANCE In this study, we demonstrate a novel hemagglutinin (HA)-specific NKp46+ NKG2A+ NK cell subset induced by influenza A virus infection. These memory NK cells show virus-specific decreased cytotoxicity and increased gamma interferon (IFN-γ) on reencountering the same influenza virus antigen. In addition, they modulate host recall responses and CD8 T cell distribution, thus bridging the innate immune and adaptive immune responses during influenza virus infection.
KEYWORDS: NK, immune memory, influenza virus, hemagglutinin, glycosylation, cytotoxicity, gamma IFN
INTRODUCTION
Influenza A virus causes epidemics and pandemics and continues to be a threat to global public health (1). Immune memory induced by vaccines or prior infections provide the most efficient protection against incoming influenza virus infection. Unfortunately, neutralizing antibody- or memory T cell-mediated recall responses can be compromised by frequent antigen drifts and antigenic shifts in influenza virus (2).
In addition to T or B cell-mediated adaptive immunity, memory-like characteristics have been found in diverse innate immune cells in the past decade (3, 4). However, memory-like innate immune cells (also called as “trained” innate immune cells) are usually equipped with restricted pathogen-recognizing patterns, which are different from the diverse receptor repertoire of adaptive immune memory. Besides the recognition of pathogen patterns, innate immune cells also develop more elaborate mechanisms to distinguish complex molecules (5, 6), which still remain largely unknow and deserve further investigation. These trained innate immune cells, including macrophages, dendritic cells (DCs), memory-like innate lymphoid cells (ILCs), and natural killer (NK) cells, generally exhibit upregulated responses on repeated stimulations (3, 7–9). However, their roles in recall responses, especially on the development and functions of adaptive immunity, wait to be illustrated.
The NK cell is a major component of innate immunity and plays a critical role in immune defense, homeostasis, and surveillance (10, 11). Based on their cytolytic capacity and cytokine production, NK cells are generally regarded as potent effector lymphocytes (12). In influenza virus infection, NK cells contribute to the restriction of virus replication and transmission, especially at the early phase of infection (13–15) through the interaction between hemagglutinin (HA) and NKp44/NKp46 (16–19). In addition to their direct antiviral effects, NK cells are capable of shaping innate and adaptive immune responses (20–23) by modulating or eliminating host immune components (24–29). However, the role of NK cells during recall immune responses remains to be clarified.
Although memory-like NK cells have been reported in humans (30–34) and experimental animal models (35–39), most of these memory-like NK cells were characterized by increased responsiveness, such as enhanced cytokine expression and cytotoxicity, upon reencounter with stimulation instead of specific antigen. Up until now, only memory-like NK cells induced by human and mouse cytomegalovirus (CMV) infections (40–43) and an HIV vaccine (44) were confirmed to be viral antigen specific. Recently, two independent studies reported the generation of human memory NK cell populations induced by seasonal influenza virus vaccination (45, 46). However, the studies failed to identify the exact antigens recognized by those memory-like NK cells. On the other hand, although CD94/NKG2C and CXCR6 were used to identify memory NK cells in some models (32, 39), the universal markers of memory NK cells are still unavailable. Moreover, it is unknown whether NK cell memory could be induced by natural influenza virus infection or whether it contributes to the initiation and regulation of recall responses.
In this study, a novel NK cell-mediated immune memory response was identified in the spleens of C57BL/6N mice with sublethal influenza virus infection. Adoptive transfer of these memory NK cells failed to rescue immunodeficient Rag2−/− γc−/− mice from challenge with the virus strain used for infecting donor mice, while sustaining protective effects against heterogenous virus infection. In contrast to other reported memory NK cell markers (30–39), these memory-like NK cells coexpressed NKp46, the marker of mature NK cells, and NKG2A, an inhibitory marker (35). While being cocultured with the same virus strain-infected DCs, these NKp46+ NKG2A+ cells lost their cytolytic activity but increased their gamma interferon (IFN-γ) response in a virus-specific manner. By using recombinant virus carrying distinct HA proteins, we further demonstrated that this NK cell memory was dependent on the recognition of N-linked glycosylation sites on influenza HA protein. The HA-specific functional switch between cytotoxicity and IFN-γ responses of memory NK cells was reversed by pifithrin-μ, an inhibitor targeting the heat shock protein 70 (HSP70)-p53 signaling pathway. In immunocompetent mice rechallenged with the same virus, splenic NKp46+ NKG2A+ NK cells were recruited to affected lung and lymphoid organs, ameliorated inflammation, and modulated the distribution of CD8+ T cells in the respiratory tract. These benefits were most apparent in mice predisposed with lung inflammation, such as a bleomycin-caused fibrosis model.
RESULTS
Prior infection impaired NK-mediated protection in a virus-specific manner.
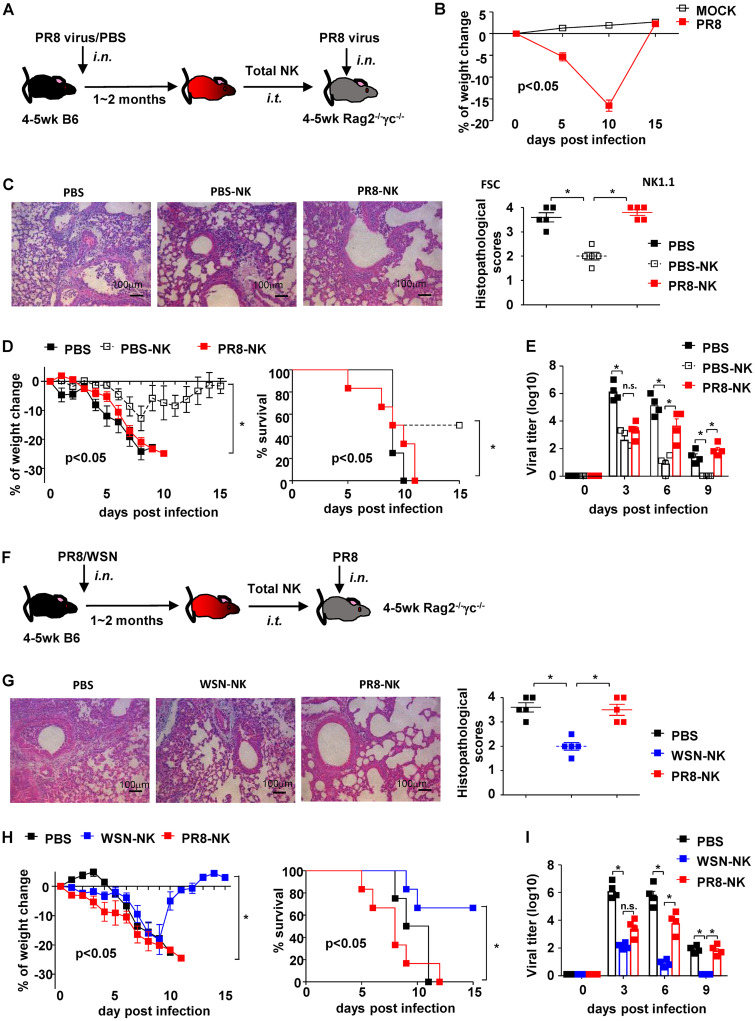
While NK cell-mediated immune responses in primary influenza virus infection are well defined, the role of NK cells during rechallenge with the same pathogen is still inconclusive. To investigate whether prior infection can modulate NK cell-mediated effects on subsequent challenge, 4- to 5-week-old C57BL/6N mice were intranasally (i.n.) inoculated with a sublethal dose of mouse-adapted influenza A virus H1N1 strain PR8 or phosphate-buffered saline (PBS) of the same volume, as a control (Fig. 1A). Most mice (>90%) showed complete recovery, manifested by regain of weight and undetected virus from the lungs of virus-infected mice by day 15 postinfection (Fig. 1B). All recovered mice were rested for two more weeks and then euthanized to isolate spleen CD3− NK1.1+ NK cells by purification with microbeads and fluorescence-activated cell sorting (FACS). Purified NK cells from PBS-treated or PR8-infected mice were adoptively transferred via the intratracheal (i.t.) route into 4- to 5-week-old Rag2−/− γc−/− mice, which lack mature T, B, and NK cells. One day later, recipient mice were challenged with a lethal dose of PR8 virus. Consistent with a previous report (47), adoptive transfer of NK cells from PBS-treated mice (PBS-NK) ameliorated weight loss of recipients, and 50% of the mice survived a lethal PR8 challenge. In contrast, NK cells from mice previously infected with PR8 virus (PR8-NK) conferred no protective effect against lethal PR8 challenge (Fig. 1C and D). Adoptive transfer of PR8-NK also delayed virus clearance in lungs of infected mice compared to that with PBS-NK treatments (Fig. 1E).
FIG 1.
Infection history impaired NK cell-mediated protection against specific virus challenge. (A) Protocol of NK isolation and adoptive transfer. (B) The weight changes of mice treated with PBS or sublethal dose of PR8 are shown; n = 12. (C) Hematoxylin and eosin staining and histopathological scores of lung tissues harvested on day 10 postinfection. (D) Weight change and survival of recipient mice. PBS-NK, NK cells isolated from PBS-treated donors; PR8-NK, NK cells isolated from PR8-infected donors. (E) Viral loads of lung tissues harvested on days 0, 3, 6, and 9 postinfection. (F) Protocol of NK isolation and adoptive transfer. (G) Hematoxylin and eosin staining and histopathological scores of lung tissues harvested on day 7 postinfection. (H) Weight change and survival of recipient mice. (I) Viral loads of lung tissues harvested on days 0, 3, 6, and 9 postinfection. WSN-NK, NK cells isolated from WSN-infected donors; PR8-NK, NK cells isolated from PR8-infected donors. Data are shown as means ± SEMs and represent 3 independent experiments, n = 4 to 6; *, P < 0.05; n.s., nonsignificant.
To investigate whether this impaired protection was virus specific, spleen NK cells isolated from C57BL/6N mice treated with H1N1 influenza virus strain WSN (WSN-NK) were adoptive transferred into Rag2−/− γc−/− mice (Fig. 1F). WSN-NK ameliorated the weight loss, improved the survival of PR8-infected Rag2−/− γc−/− mice, and attenuated viral load in affected lungs (Fig. 1G to I), which suggested that the loss of NK cell-mediated protection was dependent on the specific virus strain used to treat donors of NK cells. Consistent with this, WSN-NK failed to protect Rag2−/− γc−/− mice from a lethal WSN infection (data not shown). Taken together, these data demonstrated that prior influenza virus infection impaired NK cell-mediated protection in a virus strain-specific manner.
Prior infection induced NK memory cells with virus-specific loss of cytotoxicity and increased IFN-γ responses.
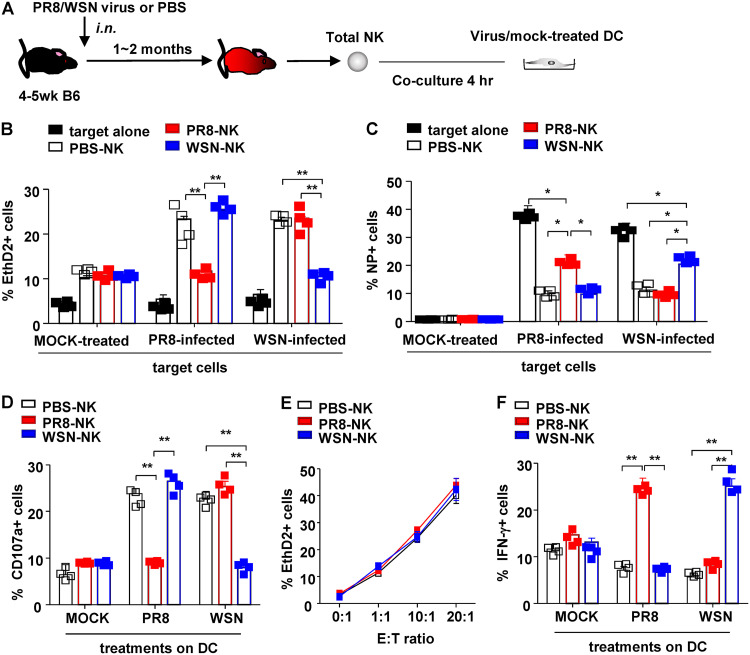
To illustrate their virus-specific function, NK cells (effector [E]) isolated from influenza virus (PR8 or WSN)- or PBS-treated mice were cocultured with live virus- or mock-treated DCs (target [T]) at an E/T ratio of 10:1 for 4 h. NK cell-mediated cytotoxicity on DCs was then determined by flow cytometry (Fig. 2A). Compared to PBS-NK, PR8-NK lost cytotoxicity on PR8-treated DCs but exhibited equivalent killing ability against WSN-treated DCs (Fig. 2B). Similarly, WSN-NK killed PR8-treated DCs but not WSN-treated DCs, resulting in higher percentage of nucleoprotein (NP)+ DCs in the culture (Fig. 2C). Consistent with their silenced virus-specific cytotoxicity, PR8-NK or WSN-NK failed to show increased CD107a expression by coculturing with PR8- or WSN-treated DCs, respectively (Fig. 2D). On the other hand, PBS-NK, PR8-NK, and WSN-NK showed comparable cytotoxicity on NK-sensitive tumor cell line YAC-1 (Fig. 2E), which suggested that loss of cytolytic ability in PR8-NK and WSN-NK was virus specific.
FIG 2.
NK cells exhibited HA-specific functions. (A) Protocol of NK isolation and in vitro coculture system. Purified NK cells (effector [E]) from different donors were cocultured with virus-treated bone marrow-derived DCs (target [T]) at an E/T ratio of 10:1 for 4 h (B, D, and E) or 1:1 for 16 h (C) at 37°C and 5% CO2. (B) NK cell-mediated cytotoxicity against distinct virus-treated DCs was determined with EthD2 staining for the last 15 min of coculture. EthD2+ DCs were regarded as apoptotic cells. (C) Expression of influenza virus NP protein in DCs. (D) Anti-CD107a monoclonal antibody was added at the start of coculture to determine the expression of CD107a on NK cells. (E) Purified NK cells (effector [E]) were then cocultured with YAC-1 cells (target [T]) at indicated E/T ratios for 4 h at 37°C and 5% CO2. NK cell-mediated cytotoxicity was determined with EthD2 staining for the last 15 min of coculture. EthD2+ YAC-1 cells were regarded as cells experiencing apoptosis; n = 4. (F) Expression of IFN-γ in NK cells was determined by intracellular staining and analyzed by flow cytometry; 10 mg/ml brefeldin A (BFA) was added into the system at the start of coculture to block the secretion of cytokines. Data are shown as means ± SEMs and represent 3 independent experiments; n = 4. *, P < 0.05;**, P < 0.01.
Unexpectedly, changes in PR8-NK- or WSN-NK-mediated cytotoxicity was not paralleled with their expressions of IFN-γ. As shown in Fig. 2F, PR8-NK showed an increase of the IFN-γ response against PR8-treated DCs but not WSN-treated DCs, while WSN-NK increased IFN-γ expression against WSN-treated DCs but not PR8-treated DCs. Taken together, these results demonstrated that NK cells isolated from influenza virus-infected donors exhibited an increased IFN-γ response but lost cytolytic capacity in a virus-specific manner.
Virus-specific memory NK cells are NKp46+ NKG2A+.
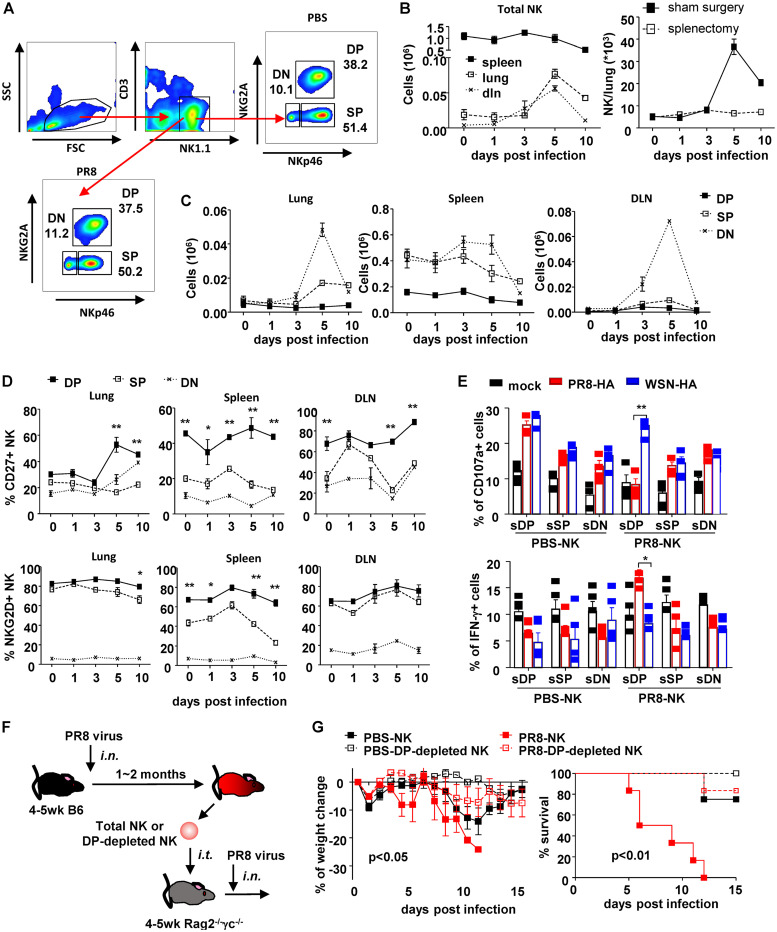
To identify the major contributor of virus-specific memory, NKp46 and NKG2A expression was used to gate total NK cells (CD3− NK1.1+) into three subsets: NKp46− NKG2A− (double negative [DN]), NKp46+ NKG2A− (single positive [SP]), and NKp46+ NKG2A+ (double positive [DP]). After complete recovery (day 30 post-primary infection), the percentages of these three NK subsets in infected mice were close to those in PBS-treated mice (Fig. 3A). Although DN represents the major expanding population in multiple organs, especially in lung and draining lymph nodes (DLN), during infection (Fig. 3B and C), only a minority of them expressed CD27 and NKG2D (Fig. 3D), markers of potent effector NK cells (12, 48). In addition, the lack of NKp46 expression in this subset suggested that this population might represent the immature phase of NK cells. Compared to SP-NK, DP-NK expressed higher levels of CD27 and NKG2D (Fig. 3D) after infection, indicating the highly activated status of this subset. Besides their expression of molecules determined by flow cytometry, functions of sorting-purified subsets were assessed in ex vivo culture. As shown in Fig. 3E, only the DP subset from PR8-NK lost CD107a expression and increased IFN-γ in the coculture with PR8-treated DCs, which suggested that NKp46 and NKG2A can be used to exclusively identify memory NK cells.
FIG 3.
NKp46+ NKG2A+ identified memory NK cell subset. (A) Flow chart of NK cell subsets identified by NKp46 and NKG2A in mock-treated mice and mice recovering from PR8 infection. DN, NKp46− NKG2A−; SP, NKp46+ NKG2A−; DP, NKp46+ NKG2A+. Dynamic changes of total NK cells (B, left) and NK subsets (C) in lung, DLN, and spleen of mice infected with sublethal dose of PR8 virus (25 μl, 103.5 TCID50), i.n., were determined by flow cytometry with counting beads. One month postinfection, some completely recovered mice received splenectomy or sham surgery. One month postsurgery, mice were infected with lethal dose of PR8 virus (25 μl, 105 TCID50), i.n. Lung-infiltrating NK cells were counted by flow cytometry at indicated times postinfection (B, right). (D) Expressions of CD27 and NKG2D in NK cell subsets in lung, DLN, and spleen during infection were determined by surface staining and flow cytometry. Difference between SP and DP populations were statistically analyzed. (E) Expressions of CD107a and IFN-γ in flow cytometry-separated NK cell subsets (sDP, sSP, and sDN) (effector [E]) were determined by surface and intracellular staining and flow cytometry after cocultured with recombinant virus-treated DCs (target [T]) at an E/T ratio of 10:1 for 4 h at 37°C and 5% CO2; n = 4. (F) Protocol of NK cells subset isolation and adoptive transfer. (G) Weight change and survival of recipient mice. DP, NKp46+ NKG2A+. Data are shown as means ± SEMs and represent 3 independent experiments; n = 4 to 6. *, P < 0.05; **, P < 0.01.
By adoptively transferring total NK cells or DP-depleted NK cells from PR8 or PBS-treated donors into Rag2−/− γc−/− mice (Fig. 3F), the in vivo effects mediated by DP subsets were compared. Consistent with results shown in Fig. 1D, PR8-NK failed to protect recipients from PR8 challenge (Fig. 3G), which could be reversed by the depletion of PR8-DP before adoptive transfer. Taken together, NKG2A+ NKp46+ NK cells demonstrated virus-specific memory under both ex vivo and in vivo conditions.
Virus-specific memory of NK cells relies on HA protein recognition.
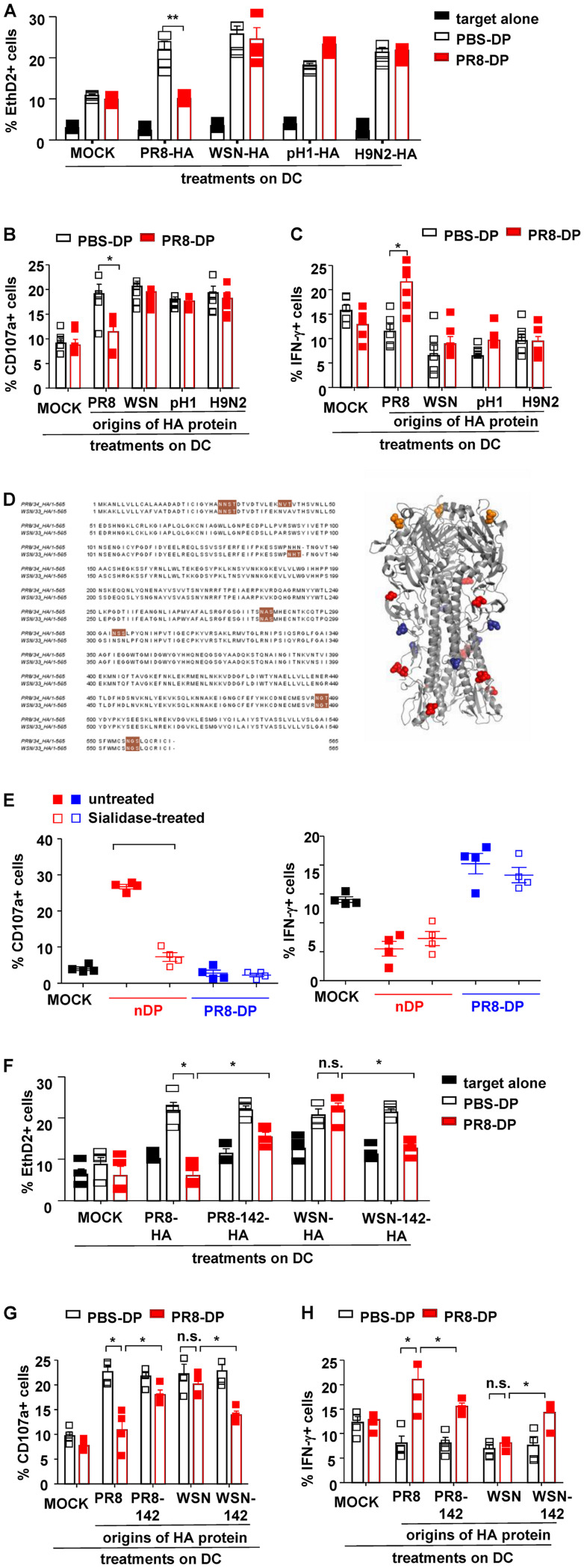
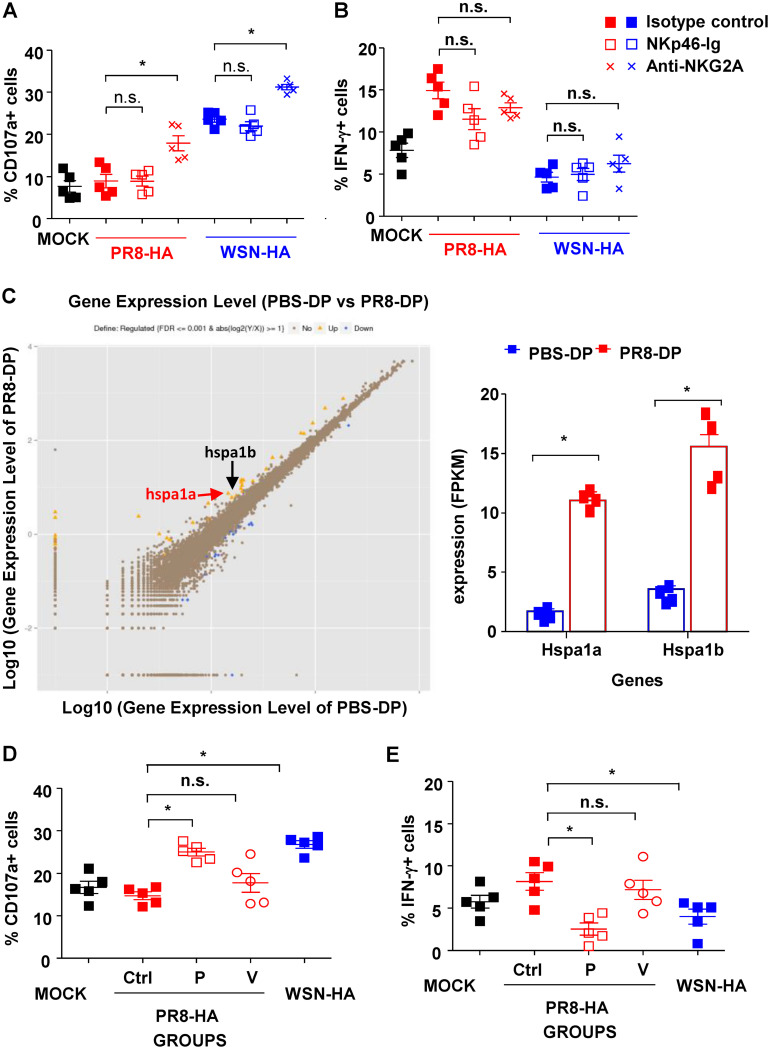
The high frequency of point mutation and antigen shift in HA proteins contributes to the epitope diversity of influenza virus (49). To determine whether distinct HA proteins could be distinguished by these virus-specific memory NK cells, HA proteins (PR8-HA, WSN-HA, pandemic H1N1-HA [pH 1-HA], or avian H9N2-HA) were expressed on recombinant PR8 virus by plasmid-based reverse genetics. DCs treated with distinct recombinant viruses were then cocultured with PBS-DP or PR8-DP as described previously (Fig. 2A). As shown in Fig. 4A, PR8-DP showed comparable cytotoxicity on recombinant virus (WSN-HA, pH1N1-HA, or H9N2-HA)-treated DCs but decreased killing capacity against PR8-HA-expressing virus-treated DCs. Consistent with their “silenced” cytolytic capacity, PR8-DP failed to increase CD107a expression in coculture with DCs treated with PR8-HA-expressing virus (Fig. 4B). On the other hand, PR8-DP cells exhibited increased IFN-γ production while coculturing with DCs treated with PR8-HA-expressing virus (Fig. 4C). These data indicated the virus-specific NK cell memory induced by influenza virus infection was dependent on HA protein recognition.
FIG 4.
NKp46+ NKG2A+ NK cells exhibited HA-specific functions. (A) NK cell-mediated cytotoxicity against distinct recombinant virus (PR8-HA, WSN-HA, pH 1-HA, or H9N2-HA)-treated DCs were determined with EthD2 staining for the last 15 min of coculture. (B) Anti-CD107a monoclonal antibody was added at the start of coculture to determine the expression of CD107a on NK cells. (C) Expression of IFN-γ in NK cells was determined by intracellular staining and analyzed by flow cytometry; 10 mg/ml BFA was added into the system at the start of coculture to block the secretion of cytokines. (D) N-Glycosylation sites are highlighted in brown. The glycosylation sites common to PR8 and WSN are colored red; those unique to PR8 are colored deep blue, and the ones unique to WSN are colored orange. (E) Purified DP-NK cells (effector [E]) from infected mice were pretreated with Arthrobacter ureafaciens sialidase for 30 min and cocultured with virus-treated bone marrow-derived DCs (target [T]) at and E/T ratio of 10:1 for 4 h at 37°C and 5% CO2. The expressions of CD107a and IFN-γ in NK were determined by surface and intracellular staining and analyzed by flow cytometry; n = 4. (F) NK cell-mediated cytotoxicity against distinct recombinant virus (PR8-HA, PR8-142-HA, WSN-HA, or WSN-142-HA)-treated DCs determined by EthD2 staining. (G and H) The expressions of CD107a and IFN-γ in NK cells determined by intracellular staining and analyzed by flow cytometry. Data are shown as means ± SEMs and represent 3 independent tests; n = 4 to 5. *, P < 0.05; n.s., nonsignificant.
HA proteins from PR8 and WSN share 90.4% identity of amino acid sequences, including five common N-linked glycosylation sites. However, PR8-HA contains two additional glycosylation sites at residues 40 and 303 (N1 numbering from ATG), while WSN-HA contains an extra glycosylation site at residue 142 (Fig. 4D). The N-linked glycosylation site at residue 142 was further investigated as it is located at the head of HA, the most potential interactive location with host cells. Importantly, sialidase treatment impaired normal NK-mediated cytotoxicity but did not reverse memory NK-mediated cytotoxicity loss or affect their IFN-γ expression (Fig. 4E). To study the role of the N-linked glycosylation site at residue 142, recombinant PR8 viruses expressing the following HA proteins were generated: PR8-HA, WSN-HA, PR8-142-HA (NHN to NHT at residues 142 to 144 of PR8-HA, thus gaining a potential N-linked glycosylation at residue 142), and WSN-142-HA (NHT to NHN at residues 142 to 144 of WSN-HA, thus losing the potential of N-linked glycosylation). As shown in Fig. 4F to H, PR8-HA-specific loss of cytotoxicity and CD107a expression in PR8-DP were partly reversed by 142 site mutation, which simultaneously deprived their IFN-γ expression. On the other hand, virus-targeted cytotoxicity and CD107a expression was purged but IFN-γ expression was enhanced in PR8-DP in coculture with DCs treated by WSN-142-HA virus. Taken together, these data suggested that the memory of NK cells was mediated by the recognition of glycosylated sites on influenza HA protein.
Pifithrin-μ treatment diminished virus-specific functions of NKp46+ NKG2A+ memory NK cells.
To determine whether NKG2A and NKp46 were involved in HA-specific function of PR8-DP cells, NKG2A and NKp46 blocking reagents were used. As shown in Fig. 5A and B, blockade of NKp46 showed no effects on the expression of CD107a and IFN-γ in PR8-DP cocultured with virus-treated DCs. Although blocking of NKG2A reversed the decreased expression of CD107a of PR8-DP while coculturing with PR8-HA-treated DCs, the treatment also increased CD107a expression in PR8-DP cocultured with WSN-HA-treated DCs (Fig. 5A). Moreover, no changes were identified in their expression of IFN-γ with NKG2A-blocking treatment (Fig. 5B). These data suggested that NKG2A- and NKp46-related signaling did not contribute to HA-specific function of memory NK cells.
FIG 5.
HSP70-regulated HA-specific NK responses. The effects of NKG2A and NKp46 blockade on HA-specific function of NKp46+ NKG2A+ NK cells (DP). Purified DP-NK cells (effector [E], with/without pretreatment with 10 mg/ml anti-NKG2A antibody or isotype control) were cocultured with DCs (target [T], with/without pretreatment with 10 mg/ml NKp46-Ig or control Ig) at an E/T ratio of 10:1 for 4 h at 37°C and 5% CO2. The expressions of CD107a (A) and IFN-γ (B) in NK were determined by surface and intracellular staining and analyzed by flow cytometry. Anti-CD107a monoclonal antibody was added to the system at the start of coculture, while 10 mg/ml BFA was added to intracellular staining at the start of coculture to block the secretion of cytokines. (C) RNA sequencing of DP-NK cells from PR8-infected mice or PBS-treated mice. (Six samples from each group were then pooled into 2 PBS-DP and 2 PR8-DP specimens for RNA extraction. The sequencing of each specimen was duplicated to make n = 4 for further analysis). (D and E) Effects of HSP70-related signal pathway blockade on HA-specific function of the DP subset; 50 mM VER-155008 or 100 mM pifithrin-m was used to pretreat DP-NK cells before coculturing with virus-treated DCs. Expressions of CD107a (D) and IFN-γ (E) in DP-NK cells were determined by surface and intracellular staining, respectively. Data are shown as means ± SEMs and represent 3 independent assays; n = 4 to 5. *, P < 0.05; n.s., nonsignificant.
To identify the molecules contributing to NK cell memory functions, gene expressions of PBS-DP and PR8-DP were determined and compared by RNA sequencing. As shown in Fig. 5C, increased expression of both hspa1a and hspa1b, critical molecules of the HSP70-related signal pathway, was found in PR8-DP compared to that in PBS-DP. More importantly, HA-specific change of CD107a and IFN-γ expression in PR8-DP was reversed by pifithrin-μ but not VER-155008 (Fig. 5D and E), while neither of these treatments modified the responses of PR8-NK to WSN-HA-treated DCs (data not shown). Although both pifithrin-μ and VER-155008 inhibit HSP70-related signals, pifithrin-μ specifically targets the p53 binding site (50). Therefore, HSP70-p53 might be a potential target in modulating NK cell memory, although the exact signaling pathway involved in its formation and function needs to be identified by further investigations.
Migration and long-term effects of NKp46+ NKG2A+ NK cells.
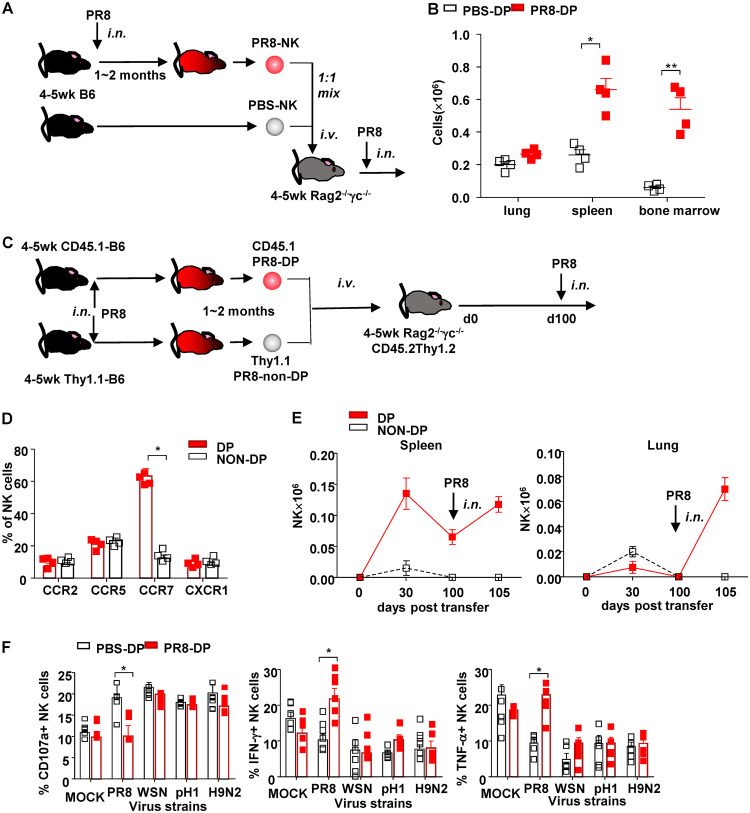
The long-term maintenance of immune memory requires an appropriate environment (“niche”) for memory cells, such as that of secondary lymphoid organs or bone marrow. To compare the migratory capacity of different NK cell subsets into spleen or bone marrow, a mixture of carboxyfluorescein succinimidyl ester (CFSE)-labeled PR8-NK and PBS-NK was adoptively transfer via tail vein injection (i.v.) to Rag2−/− γc−/− mice immediately prior to infection (Fig. 6A). Compared to PBS-DP, more PR8-DP were found to accumulate in spleen and bone marrow of recipients on day 10 postinfection (Fig. 6B), which might benefit their long-term survival (51).
FIG 6.
NKp46+ NKG2A+ NK cells maintained long-term virus-specific memory. (A) Protocol of in vivo trafficking of NK cells and NK subsets. (B) Distribution of NK cell subsets in lung, spleen, and bone marrow of recipients on day 10 postinfection. (C) Protocol of long-term trafficking of NK subsets. (D) Chemokine receptors of DP and non-DP NK cells. (E) Quantity of DP (CD45.1+) and non-DP (Thy1.1+) NK cells in spleen and lung at indicated times posttransfer. (F) Four- to 5-week-old C57BL/6N mice were treated with sublethal dose of PR8 (25 μl, 103.5 TCID50) or PBS of the same volume, i.n. One year later, completely recovered mice were sacrificed, and spleen NK cells were purified by a magnetic bead isolation kit followed with flow cytometry sorting. Purified NK cells (effector [E]) were then cocultured with virus-treated bone marrow-derived DCs (target [T]) at an E/T ratio of 10:1 for 4 h at 37°C and 5% CO2. The expressions of CD107a, IFN-γ, and TNF-α in NK cells were determined by surface and intracellular staining and analyzed by flow cytometry. Anti-CD107a monoclonal antibody was added to the system at the start of coculture, while 10 mg/ml BFA was added to intracellular staining at the start of coculture to block the secretion of cytokines. Data are shown as means ± SEMs and represent 3 independent experiments; n = 4 to 5. *, P < 0.05; **, P < 0.01.
To track the long-term dynamics of virus-specific memory NK cells, PR8-DP NK cells from CD45.1 mice and PR8-non-DP NK cells from Thy1.1 mice were harvested and mixed before transfusion into Rag2−/− γc−/− mice (Fig. 6C). Consistent with their higher expression of CCR7 (Fig. 6D), DP-NK exhibited a higher capability to migrate into spleen than non-DP and maintained their survival until 100 days post-adoptive transfer (Fig. 6E). More importantly, these long-lived DP NK cells were able to be expanded on day 5 postinfection and recruited to lungs of recipients (Fig. 6E). Meanwhile, virus-specific CD107a and IFN-γ expression changes were identified in DP-NK cells 1 year after primary PR8 virus infection (Fig. 6F). Taken together, these data demonstrated a long-term persistence of NK cell memory in influenza virus-infected mice.
Virus-specific memory NK cells modulated recall responses.
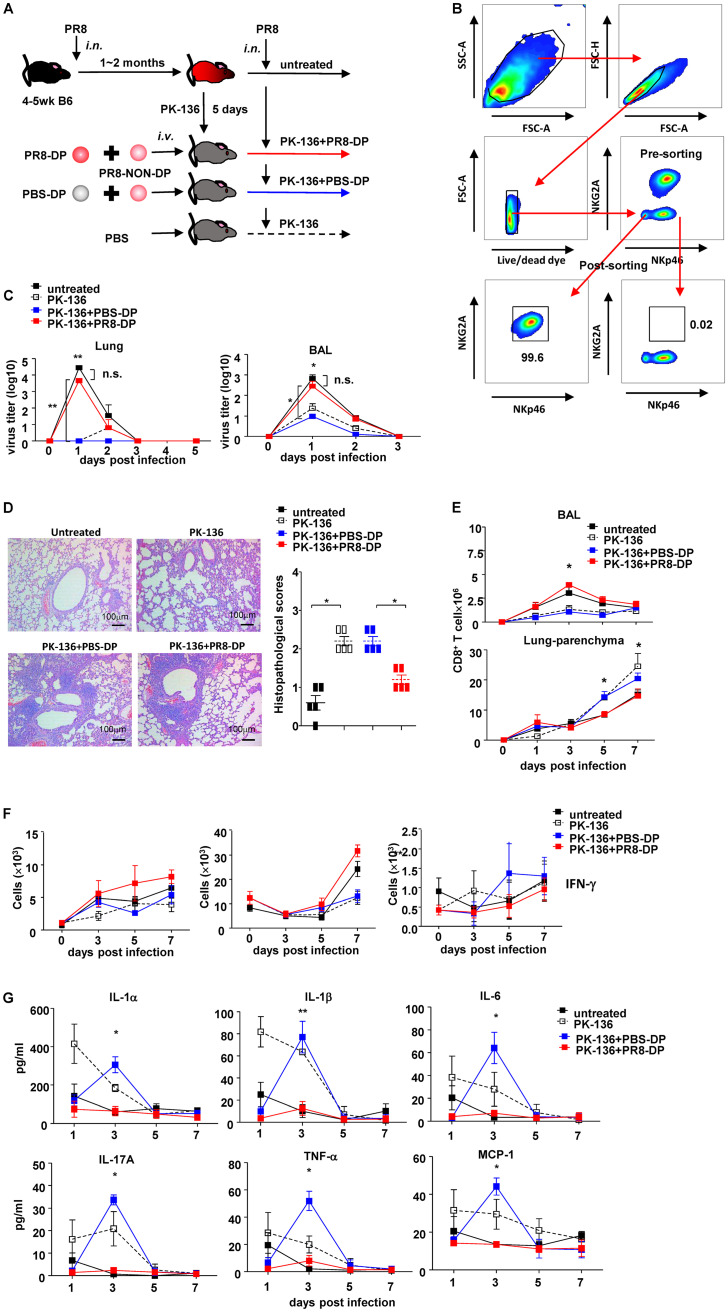
To further clarify the role of virus-specific memory DP-NK during host recall responses, an NK replacement model was established as shown in Fig. 7A and B. Briefly, recipient mice were treated with PK-136 monoclonal antibody (MAb) to deplete NK cells after complete recovery from sublethal PR8 infection. On day 5 posttreatment, more than 99.6% of peripheral NK cells were cleared, and the deprivation effects were sustained for more than 14 days (21). Since splenic NK cells represent the majority of lung-infiltrating lymphocytes during the first week postinfection (Fig. 3B), NK cell subsets isolated from spleens of PBS- or sublethal PR8-treated mice were then transferred into NK cell-depleted recipients via tail vein (i.v.) immediately prior to challenge of a lethal dose of PR8. Although mice from all 4 groups exhibited complete resistance to reinfection and lost no weight, adoptive transfer of PBS-DP exhibited higher efficiency on virus clearance (determined by viral load in both homogenized lung tissues and bronchoalveolar lavage [BAL] fluid) than PR8-DP on day 1 postinfection (Fig. 7C). Despite an asymptomatic course, histological examination of lung tissues harvested on day 7 postinfection showed injury and infiltration of leukocytes in all groups (Fig. 7D). However, compared to diffuse interstitial pneumonia in the PK-136 alone group and PK-136 plus PBS-DP (PK-136+PBS-DP) group, untreated and PK-136 plus PR8-NK (PK-136+PR8-NK) groups exhibited restricted inflammation around bronchus (Fig. 7D). CD8+ T cells are major lung-infiltrating leukocytes during the recall response, and their distribution correlated with the outcome of disease. In untreated and PK-136+PR8-DP groups, CD8+ T cells showed increased recruitment in BAL fluid on day 3 postinfection, whereas PK-136 alone and PK-136+PBS-DP treatment resulted in massive infiltration of CD8+ T cells in parenchyma of lungs on day 5 and 7 postinfection (Fig. 7E). Correspondingly, PK-136+PR8-DP treatment increased the absolute number of splenic IFN-γ+ CD8+ T cells (Fig. 7F), although the interaction between memory NK cells and CD8+ T cells needs to be further investigated. This, accompanied with lower concentrations of interleukin 1α (IL-1α), IL-1β, IL-6, IL-17A, tumor necrosis factor α (TNF-α), and monocyte chemoattractant protein 1 (MCP-1) in affected lungs of mice from the PK-136+PR8-DP group (Fig. 7G), suggested that PR8-DP modified the distribution of infiltrating leukocytes and ameliorated interstitial inflammation in lung during the recall response. On the other hand, very low levels of IFN-γ were detected in homogenized lung tissues from recipient mice (data not shown), which might be due to the small number of adoptive transferred NK cells.
FIG 7.
NKp46+ NKG2A+ NK cells modulated recall responses. (A) Protocol of NK cell isolation and adoptive transfer. untreated, NK-competent mice; PK-136, NK-depleted mice; PBS-DP, NK-depleted mice receiving mixture of DP subset from PBS-treated donors and non-DP subsets from PR8-infected donors; PR8-DP, NK-depleted mice receiving NK cells isolated from PR8-infected donors. (B) Pre- and postsorting flow plots of DP-NK and non-DP-NK purification. (C) Virus titers in homogenized lung tissue and BAL fluid of recipient mice post-second infection determined by TCID assay. (D) Hematoxylin and eosin staining and histopathological scores of lung tissues harvested on day 7 post-second infection; n = 5. (E) Dynamic changes of CD8 in BAL fluid and lung parenchyma of mice infected with lethal dose of PR8 virus (25 μl, 105 TCID50), i.n., after depletion of NK cells and adoptive transfer of PBS-DP or PR8-DP. (F) Memory NK subset increased IFN-γ expression in spleen CD8+ T cells. Four- to 5-week-old C57BL/6N mice were treated with sublethal dose of PR8 (25 μl, 103.5 TCID50) or PBS of the same volume, i.n. One to 2 months later, completely recovered mice were sacrificed, and spleen NK cell subsets were purified by a magnetic bead isolation kit followed with flow cytometry sorting. Purified DP subset from PBS-treated donors (PBS-DP) or PR8-infected donors (PR8-DP) were adoptively transferred with non-DP cells from PR8-infected mice into 5- to 6-week-old NK-depleted (by PK-136) C57BL/6N mice, i.v., at 0.25 × 106 cells/mouse in 0.2 ml PBS. Recipient mice were then infected with a lethal dose of PR8 virus (25 μl, 105 TCID50), i.n., 4 h after cell transfusion. Expressions of IFN-γ of lung, DLN, and spleen CD8+ T cells of recipient mice are shown; n = 4. (G) Concentrations of IL-1α, IL-1β, IL-6, IL-17A, TNF-α, and MCP-1 in lung were determined by flow cytometry; n = 4. Data shown as means ± SEMs and represent 3 independent experiments. *, P < 0.05; **, P < 0.01; n.s., nonsignificant.
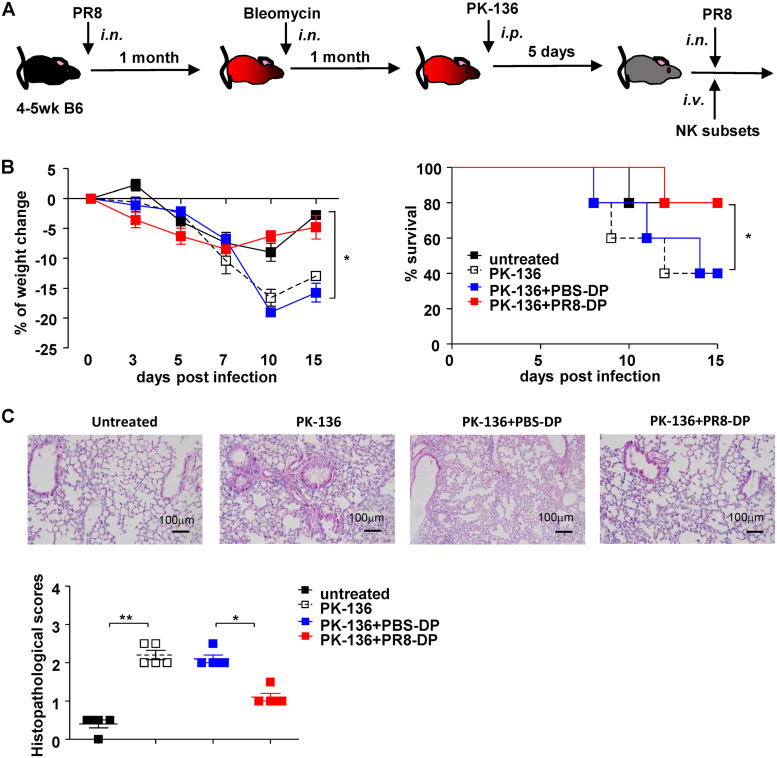
To further confirm the effect of PR8-DP on lung inflammation during the recall response, we established a lung inflammation model in PR8-infected mice with bleomycin treatment (Fig. 8A). As shown in Fig. 8B and C, PR8-DP transfer significantly improved the weight change, survival, and the pathology in the lungs of recipient mice. Taken together, these data showed that memory NK cells modulated the distribution of CD8+ T cells during recall responses and subsequently benefit the outcome of disease, especially in mice predisposed with lung inflammation.
FIG 8.
NKp46+ NKG2A+ NK cells ameliorated PR8 infection in mice with lung fibrosis. (A) Protocol of NK cell adoptive transfer in mice suffering lung inflammation caused by bleomycin. (B) Weight change and survival of mice. Data shown as means ± SEMs and represent 3 independent experiments; n = 5. (C) Hematoxylin and eosin staining and histopathological scores of a bleomycin-induced lung inflammation model treated with different NK cell subsets; n = 5. Data shown as means ± SEMs and represent 3 independent experiments. *, P < 0.05; **, P < 0.01.
DISCUSSION
NK cells are mobilized to restrict invasion of potential pathogens at very early phase of infection. This protection could be accomplished through production of cytokine and/or direct killing of virus-infected target cells. Although prior exposure to pathogens is unnecessary for NK cell-mediated immune responses, preactivation of NK cells with proinflammatory cytokines was reported to enhance their functions (40–42). With the blurring of divisions between innate and adaptive immunity, the “trained” profiles of some innate immune cells are now defined as a subtype of immune memory (52). However, only a minor fraction of these “trained memory” cells was confirmed to be antigen specific, such as m157-specific Ly49H+ NK cells induced by mouse cytomegalovirus (MCMV) infection (37, 38, 46). In this study, an NKG2A+ NKp46+ NK cell subset was found to possess influenza virus HA-specific memory (Fig. 3) and modify the recall responses against the same pathogens (Fig. 7).
Distinct from T cells and B cells, NK cells lack Rag-mediated recombinant receptors for recognition of variant antigens. However, NK cells are equipped with multiple receptor families, such as NK cell receptors (NCRs), killer cell inhibitory receptors (KIRs), and killer cell lectin-like receptors (KLRs) (53, 54). Recently, donor and recipient HLA-KIR matching has been applied to predict the outcomes of human transplantation (55). Similarly, moderately expanded and diversified NKG2 genes were found in macaques, which might represent one possible platform used by NK cells to distinguish diverse stimulants (56). In addition, human 2B4 and NTB-A receptors were also found to bind influenza virus HA (57); whether they might contribute to human NK cell memory deserves further investigation.
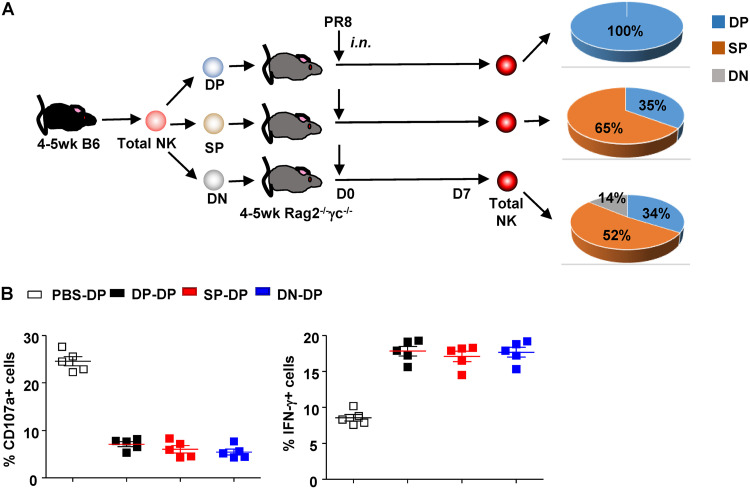
Distinct from liver memory NK cells reported recently (36), splenic NK cells represent the major origin of lung-infiltrating NK cells (Fig. 3B), which enhances their contribution during the recall response against influenza infection, although the phenotypes and functions of lung-resident NK cells (58) during recall responses deserve further investigations. Consistent with that, the lung DP-NK subset also showed HA-specific memory functions (data not shown). To clarify the development of DP-NK, purified splenic NK subsets (DN, SP, and DP) from mock-treated mice were adoptively transfer into Rag2−/− γc−/− mice prior to infection. As shown in Fig. 9A, both DN and SP differentiated into DP, while transferred DP maintained their phenotypes, suggesting a highly differentiated status of DP. In addition, DP derived from different subsets exhibited comparable virus-specific function when culturing with PR8-treated DCs (Fig. 9B).
FIG 9.
Differentiation and function of transferred NK cell subsets in recipient mice. (A) Percentages of NK subsets in recipient mice receiving DP-, SP-, or DN-NK. (B) DP-NK from different recipients (DP-DP, DP isolated from mice receiving DP-NK; SP-DP, DP isolated from mice receiving SP-NK; DN-DP, DP isolated from mice receiving DN-NK; PBS-DP, DP isolated from naive C57BL/6N mice) were purified by flow sorting and cocultured with PR8-treated bone marrow-derived DCs (target [T]) at an E/T ratio of 10:1 for 4 h at 37°C and 5% CO2. Expressions of CD107a and IFN-γ in NK were determined by surface and intracellular staining and analyzed by flow cytometry. Anti-CD107a monoclonal antibody was added to the system at the start of coculture, while 10 mg/ml BFA was added to intracellular staining at the start of coculture to block the secretion of cytokines; n = 5. Data are shown as means ± SEMs and represent 2 independent experiments.
On the other hand, the induction of NK cell memory relied on the severity of primary infection. In our preliminary experiments, mild (<10% weight loss) infection failed to generate virus-specific memory NK cells, which might have resulted from the extent of virus replication and levels of proinflammatory factors such as IFN-α/β, IL-12, IL-15, and IL-18 (30, 34, 42, 59–62). However, the blockade of IFN-α/β, IL-1β, IL-12, and IL-18 in this system failed to abolish the memory function of DP-NK cells (data not shown). In contrast to chronic infections, such as CMV and HIV infections, where persistence of the infecting virus makes it difficult to exclude the possibility that an observed “memory-like” phenotype is driven by antigen persistence (33, 37, 63), influenza virus causes an acute infection associated with complete clearance of influenza virus within 15 days postinfection. Therefore, persistence of DP-NK memory is independent of continuous pathogen stimulation. The factors contributing to the generation and maintenance of memory NK cells deserves further investigation.
Rather than simply upregulating their responsiveness, preactivation might modulate the function of NK cells at multiple levels (48), such as the differentiation of human NK cells from CD56hi CD16− into CD56int CD16+ cells, which was accompanied with the decrease of cytokine production and increase of cytolytic ability (64). Similarly, NK cells isolated from influenza virus-infected mice lost their cytotoxicity but upregulated their IFN-γ production in an HA-specific manner (Fig. 4). This functional switch led to longer persistence of virus in lung, leading to a disastrous outcome for immunodeficient Rag2−/− γc−/− mice receiving PR8-NK (Fig. 1). In immunocompetent hosts, decreased cytotoxicity was well compensated by robust expansion and accumulation of CD8+ T cells and thus did not affect the overall morbidity (e.g., weight loss) and survival. On the other hand, NK cell-mediated cytotoxicity also contributes to immunopathology and might aggravate diseases in patients predisposed with inflammatory diseases such as chronic obstructive pulmonary disease (COPD) and the aged. Under these conditions, virus-specific silenced cytotoxicity of memory NK cells helps avoid the excessive inflammation and results in an improved outcome, as shown in Fig. 8B and C. Moreover, the accumulation of CD8+ T cells in BAL fluid instead of interstitial homogenate (Fig. 7E) with the help from memory NK cells benefits the virus clearance and further restricts lung inflammation (Fig. 7D).
Although memory NK cells expressed NKG2A and NKp46 (Fig. 3 and 6), neither NKG2A- (65) nor NKp46-related signals were required for virus-specific function of memory NK cells (Fig. 5A and B). Similarly, sialidase treatment impaired normal NK-mediated cytotoxicity but did not reverse memory NK-mediated cytotoxicity loss or affect their IFN-γ expression (Fig. 4E). Subsequent blocking assays indicated the critical roles of HSP70 and p53 in the HA-specific function switch in memory NK cells (Fig. 5D and E). HSP70, together with IL-15, heat shock cognate protein 70 (HSC70), Toll-like receptor 4 (TLR4), and CD91, was recently shown to be essential for NK cell-mediated tolerance at decidua basalis during pregnancy (66). In another report, HSP70 was also found to directly respond to HA-induced signals although details remained to be clarified (67). p53 expression could be upregulated by influenza virus infection and contribute to viral clearance and thus represents a target by some viruses to facilitate their replication and spreading (68). Clarifying the roles of HSP70 and p53 during influenza virus infection might boost novel antiviral strategies. The involvement of other genes in NK cell memory could not be excluded due to the relatively small number of samples used for sequencing in this study and the single time point of sample collection.
In summary, HA-specific NKp46+ NKG2A+ memory NK cells bridge innate and adaptive immunity and thus modulate the recall responses on rechallenge of influenza virus through a function switch from cytolytic cells into cytokine-expressing effectors.
MATERIALS AND METHODS
Animals.
Rag2−/− γc−/− mice were purchased from Taconic and maintained in an individual ventilated cage (IVC) system in a specific-pathogen-free (SPF) environment in the laboratory animal unit, the University of Hong Kong. C57BL/6N and BALB/c mice were purchased from the laboratory animal unit, the University of Hong Kong. CD45.1-B6 and Thy1.1-B6 mice were purchased from Jackson Laboratory. All manipulations on animals were performed in compliance with the Animals (Scientific Procedures) Act, 1986 (UK) (amended in 2013) and approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR), Hong Kong (approval number CULATR 3283-14). All sections of this report adhered to the ARRIVE guidelines for reporting animal research.
Viruses and animal models.
Mouse-adaptive influenza virus PR8 (A/Puerto Rico/8/1934 H1N1), influenza virus WSN (A/WSN/1933 H1N1), seasonal influenza virus (A/HK/54/98 H1N1), pandemic influenza virus (A/California/7/09 H1N1), and avian influenza virus (A/quail/HK/G1/97 H9N2) were cultured in MDCK (Madin-Darby canine kidney cell line). Viral titer was determined by daily observation of cytopathic effect in MDCK infected with serial dilutions of virus stock; median tissue culture infective dose (TCID50) was calculated according to the Reed-Muench formula. For primary infection, 4- to 6-week-old female C57BL/6N mice were infected with a sublethal dose of PR8 or WSN virus (25 μl, 103.5 TCID50 or 103 TCID50, respectively), i.n. For the second challenge, completely recovered mice were infected with a lethal dose of PR8 or WSN virus (25 μl, 105 TCID50 or 104 TCID50, respectively), i.n. For adoptive transfer experiments, 4- to 6-week-old Rag2−/− γc−/− mice were infected with a lethal dose of PR8 or WSN virus (25 μl, 103 TCID50), i.n. Three to 6 mice per group were used for each independent experiment, and all experiments were repeated 2 times to obtain unbiased data, without unnecessary overuse of experimental animals. Mice with >25% weight loss were sacrificed, counted as death. Bleomycin-induced lung inflammation model was induced by injection with bleomycin (Nippon Kayaku Co. Ltd.) at an intratracheal dose of 1 mg of bleomycin per g of body weight after anesthesia.
In all animal experiments, mice were monitored twice a day during the whole experiment, while food and water were provided ad libitum. Soft and clean bedding, a quiet environment, and circadian light were provided to reduce animal stress. No unexpected death occurred during this study, and mice were euthanized by cervical dislocation under anesthesia (by intraperitoneal [i.p.] injection of ketamine plus xylazine at the final concentrations of 7.5 mg/kg and 0.88 mg/kg, respectively) when the study was completed or when one of following conditions was observed: weight loss of >30%; body temperature change of >2°C; cardiac/respiratory rate change of >50%; signs of severe pneumonia (very weak and precomatose). To minimize animal suffering and distress, all invasive manipulations were carried out under anesthesia as described above.
Depletion of mouse NK cells.
To produce PK-136 antibodies, 4- to 6-week-old BALB/c mice were pretreated with 0.5 ml incomplete Freud’s adjuvant (IFA), i.p., and then injected with PK-136 hybridoma cells (ATCC), 5 × 106 cells/mouse, i.p. Two weeks later, ascites was harvested and purified with protein G columns. To deplete NK cells, 0.2 mg purified PK-136 or anti-GM1 antibody (Asialo) diluted in 0.5 ml PBS was injected into each mouse i.p.
Adoptive transfer of mouse NK cells.
Mouse NK cells were isolated from spleen cells with a mouse NK isolation kit II (Miltenyi microbeads) and stained with fluorescence-labeled antibodies. Total NK cells or NK cell subsets were then sorted by flow cytometry (FACSAria; BD). For rescue experiments, total NK cells or NK subsets were injected, i.t., into Rag2−/− γc−/− mice, 0.2 × 106 cells/mouse in 0.1 ml PBS. For recall response experiments, NK subsets were injected into recipient mice, i.v., 0.25 × 106 cells/mouse in 0.2 ml PBS.
Trafficking assays.
Mouse NK cells were isolated from completely recovered mice pretreated with a sublethal dose of PR8 virus or PBS of the same volume and purified with a mouse NK isolation kit II (Miltenyi microbeads). Purified NK cells from PR8- or PBS-treated mice were stained with carboxyfluorescein succinimidyl ester (CFSE) or eFluor405-CFSE, respectively. Labeled NK cells were then mixed at the ratio of 1:1 and injected into 4 -to 6-week-old female Rag2−/− γc−/− mice, i.v., 0.2 × 106 cells/mouse in 0.2 ml PBS.
Bronchoalveolar lavage fluid harvest.
The thoracic cage and neck of the mouse were exposed, and a small incision was made in the trachea. A 23-gauge needle connected to a tube was placed, and then 1 ml saline was injected into the animal’s lungs and aspirated. Four wash procedures were carried out in each animal. The collected aspirations from each animal were pooled for further analysis.
Immunohistochemistry assays of lungs.
The lungs from mice were harvested at the designated time points postinfection, fixed with 10% formalin, and maintained in 95% ethanol. Paraffin-embedded lung sections were prepared according to standard protocols and stained with hematoxylin and eosin. All lung sections were screened, and five fields from each sample were selected randomly by 2 independent pathologists for evaluating the levels of pathology among different groups. The following criteria were used for scoring edema, hyaline membrane formation, and necrotic cellular debris: 0, none; 1, uncommon detection in <5% of lung fields (×200 magnification); 2, detectable in up to 33% of lung fields; 3, detectable in up to 33% to 66% of lung fields; 4, detectable in >66% of lung fields.
Determination of virus copy and inflammatory cytokines/chemokines in lungs.
The lungs from infected mice were harvested at designated time points postinfection and homogenized in 2 ml of RPMI 1640 medium. After centrifugation at 1,500 × g for 15 min, the supernatants were collected for determining viral titer (TCID50) as described above or concentrations of proinflammatory cytokines and chemokines with murine cytokine and chemokine assay kits (Bender MedSystems).
Preparation of recombinant virus.
The recombinant viruses were generated using the eight-plasmid system as described previously (69). HA genes of A/California/4/09 virus and the avian influenza virus A/Quail/Hong Kong/G1/97 (H9N2) were amplified and cloned into the pHW2000 vector as described. Plasmids encoding the eight genes of A/PR/8/34 (H1N1) (pHW191-PB2, pHW192-PB1, pHW193-PA, pHW194-HA, pHW195-NP, pHW196-NA, pHW197-M, and pHW198-NS) and HA genes of A/WSN/33 (H1N1) (pHW184-HA) were kindly provided by Robert Webster at St. Jude Children’s Research Hospital. The addition of the N-linked glycosylation site at residue 142 of PR8-HA (PR8-142-HA, with changes from NHN to NHT at residues 142 to 144) and the removal of the N-linked glycosylation site at residue 142 of WSN-HA (WSN-142-HA, with changes from NHT to NHN at residues 142 to 144) were achieved by site-directed mutagenesis. Recombinant viruses with seven internal genes (PB2, PB1, PA, NP, NA, M, and NS) derived from the A/PR/8/34 virus and different HA genes derived from A/California/4/09 (pdmH1N1), A/Quail/Hong Kong/G1/97 (H9N2), A/PR/8/34 (H1N1), A/WSN/33 (H1N1), PR8-142-HA (H1N1), or WSN-142-HA (H1N1) were generated by cotransfecting plasmids in subconfluent human embryonic kidney 293T cells (TransIT-LT1; Mirus). The rescued viruses were further propagated in MDCK cells for two or three passages at a multiplicity of infection (MOI) of 0.001 PFU/cell. The HA genes of the recombinant viruses were verified by Sanger sequencing.
Generation of bone marrow DCs.
Mouse bone marrow cells were harvested from the femur and cocultured in 10% fetal bovine serum (FBS)-RPMI 1640 medium supplemented with recombinant 30 ng/ml IL-4 and 20 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF; Peprotech) for 5 days. To induce the maturation of DCs, 0.1 μg/ml lipopolysaccharide (LPS; Invitrogen) was added to the culture for 16 h at 37°C and 5% CO2.
In vitro coculture system.
Mouse bone marrow-derived DCs pretreated with mock virus, wild-type virus, or recombinant virus at an MOI of 2 for 16 h were harvested as target cells (T) and then cocultured with mouse spleen NK cells or NK subsets (effector [E]) in 10% FBS-RPMI 1640 medium at and E/T ratio of 10:1 for 4 h (for cytotoxicity and CD107a/IFN-γ expression assays) or 1:1 for 16 h (for NP staining assay), in 37°C and 5% CO2. The expressions of cell surface or intracellular molecules in NK cells or DCs were determined with FACS LSR-II (BD) and FlowJo software (Tree Star).
Cytotoxic assay.
The YAC-1 cell line or DCs (target [T]) were stained with CFSE and cocultured with mouse NK cells (effector [E]) at different E/T ratios for 4 h. Cells were then stained with ethidium homodimer-2 (EthD2) (Thermo Fisher) to identify apoptotic targets and analyzed with FACS LSR-II (BD) and FlowJo software (Tree Star).
Quantification of viral copies by reverse transcription-PCR.
The homogenized lung tissues were harvested for extraction of total RNA by TRIzol LS reagent according to the manufacturer’s instructions (Invitrogen). The cDNA was synthesized with oligo(dT)12−18 primers and Superscript II reverse transcriptase (Invitrogen). Viral matrix gene copies were quantified by SYBR green fluorescence after a real-time PCR procedure (forward primer, 5′-CTTCTAACCGAGGTCGAAACG-3′; reverse primer, 5′-GGCATTTTGGACAAAGCGTCTA-3′) on a PRISM 7900 sequence detection system (Applied Biosystems).
Quality analysis and mapping of RNA expression.
Total RNA of distinct NK subsets was isolated by using a Qiagen RNA isolation kit and treated with RNase-free DNase I (New England BioLabs, MA, USA), to remove contaminating genomic DNA. The cDNAs were then fragmented by nebulization followed by the standard Illumina protocol to create the mRNA sequencing (mRNA-seq) library. Clean reads were obtained by removing reads containing adapter or poly(N) and low-quality reads. All the downstream analyses were based on good-quality clean reads and mapped to mouse genome (ftp://ftp.ensembl.org/pub/release-81/gtf/mus_musculus/). Briefly, 75 FastQ single reads (n = 23.6 million average per sample) were trimmed using Trimmomatic (v 0.33) enabled with the optional “-q” option; 3-bp sliding-window trimming from the 3′ end requiring minimum Q30. Quality control on raw sequence data for each sample was performed with FastQC. Read mapping was performed via TopHat v2.0.9 and Cufflinks v2.1.1. using the mouse genome (mm10) as a reference. Gene quantification was performed via Feature Counts for raw read counts. Differentially expressed genes were identified using the edgeR (negative binomial) feature in CLCGWB (Qiagen, Redwood city, CA) using raw read counts. We filtered the generated list based on a minimum 2× absolute fold change and false-discovery rate (FDR) corrected P value of <0.05.
Flow cytometric assays and analysis.
Cells were stained for surface markers with the following antibodies: anti-CD3 (145-2C11; BioLegend), anti-CD8 (53-6.7; BioLegend), anti-NK1.1 (PK-136; BioLegend), anti-NKp46 (29A1.4; BioLegend), anti-NKG2A (16A11; BioLegend), anti-NKG2D (CX5; BioLegend), anti-CD11b (M1/70; BioLegend), anti-CD25 (PC61.5; eBioscience), anti-CD27 (LG.3A10; BioLegend), anti-CD69 (H1.2F3; BioLegend), anti-CD107a (1D4B; BioLegend), and anti-TRAIL (N2B2; BioLegend) antibodies. For intracellular staining, cells were fixed, permeabilized, and then stained with anti-IFN-γ (XMG1.2; BioLegend), anti-TNF-α (MP6-XT22; BioLegend), and anti-influenza virus NP protein (C43; Abcam) antibodies or their relevant isotype controls as described previously (70, 71). For cell counting assays, CountBright absolute counting beads (Invitrogen) were added. Single cell gating and a LIVE/DEAD cell viability assay kit (Invitrogen) were used to exclude dead cells and aggregated cells before analysis and cell sorting. All data were acquired on a FACS LSR-II (BD) and analyzed by FlowJo software (Tree Star).
Blocking assay.
To block the NKG2A-related signal pathway, 10 mg/ml anti-NKG2A (bs-2411R; Bioss) and its isotype control were added to NK cells for 1 h prior to the initiation of coculture. To block the NKp46-related signal pathway, 10 mg/ml NKp46-Fc chimera protein (R&D) and control Ig-Fc protein were added to mock or virus-treated DC for 1 h prior to the initiation of coculture. For blocking assays, 50 μM VER-155008 or 100 μM pifithrin-μ (Sigma-Aldrich) was added, respectively, to the in vitro coculture system at the start of coculture.
Sialidase treatment.
To determine whether the HA binding of NK cells was dependent on sialic acid, NK cell subpopulations were treated with Arthrobacter ureafaciens sialidase (Sigma) for 30 min and then extensively washed prior to coculture as previously described (16).
Statistical analysis.
Data are presented as means ± standard errors of the means (SEMs). Multiple regression analysis was used to test the differences in the body weight changes between different groups adjusted for time after infection. The differences in cytotoxicity and virus copy of in vitro experiments and viral load or concentrations of proinflammatory cytokines/chemokines were analyzed by one-way analysis of variance (ANOVA) and unpaired two-tailed Student’s t test. The difference for survival and weight change was determined by Kaplan-Meier log-rank test. All analyses were accomplished on GraphPad Prism, and a P value of <0.05 was considered to be significant.
Data availability.
Raw data files have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE124321.
ACKNOWLEDGMENTS
We thank Robert Webster (St. Jude Children’s Research Hospital) for kindly providing plasmids encoding the eight genes of A/PR/8/34 (H1N1) (pHW191-PB2, pHW192-PB1, pHW193-PA, pHW194-HA, pHW195-NP, pHW196-NA, pHW197-M, and pHW198-NS) and HA genes of A/WSN/33 (H1N1) (pHW184-HA and pHW186-NA).
This work was supported in part by the General Research Fund, Research Grants Council of Hong Kong (17121214, 17115015, 17126317, 17122519), and the Theme-based Research Scheme from the Research Grants Council of the Hong Kong SAR, China (project number T11-705/14N).
J.Z. and W.T., initiated and designed the study. J.Z., L.W., H.-L.Y., M.L., Y.L., O.T., W.-F.W., N.K., K.-T.L., C.H., and J.Y. performed experiments and collected data. J.Z. and L.W. analyzed data. J.Z., L.W., H.-L.Y., S.P., W.T., and M.P. drafted the article. J.Z., W.T., M.P., and Y.-L.L. supervised the study. All authors contributed to review and revision and have seen and approved the final version.
We declare no competing interests.
Contributor Information
Jian Zheng, Email: jian-zheng-1@uiowa.edu.
Malik Peiris, Email: malik@hku.hk.
Wenwei Tu, Email: wwtu@hku.hk.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCES
- 1.Medina RA, Garcia-Sastre A. 2011. Influenza A viruses: new research developments. Nat Rev Microbiol 9:590–603. 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, Garcia-Sastre A. 2018. Influenza. Nat Rev Dis Primers 4:3. 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapp M, Wiedemann GM, Sun JC. 2018. Memory responses of innate lymphocytes and parallels with T cells. Semin Immunopathol 40:343–355. 10.1007/s00281-018-0686-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamon MA, Quintin J. 2016. Innate immune memory in mammals. Semin Immunol 28:351–358. 10.1016/j.smim.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Rouzaire P, Luci C, Blasco E, Bienvenu J, Walzer T, Nicolas JF, Hennino A. 2012. Natural killer cells and T cells induce different types of skin reactions during recall responses to haptens. Eur J Immunol 42:80–88. 10.1002/eji.201141820. [DOI] [PubMed] [Google Scholar]
- 6.Rydyznski CE, Waggoner SN. 2015. Boosting vaccine efficacy the natural (killer) way. Trends Immunol 36:536–546. 10.1016/j.it.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitrovic M, Arapovic J, Traven L, Krmpotic A, Jonjic S. 2012. Innate immunity regulates adaptive immune response: lessons learned from studying the interplay between NK and CD8+ T cells during MCMV infection. Med Microbiol Immunol 201:487–495. 10.1007/s00430-012-0263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardiner CM, Mills KH. 2016. The cells that mediate innate immune memory and their functional significance in inflammatory and infectious diseases. Semin Immunol 28:343–350. 10.1016/j.smim.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Sohrabi Y, Godfrey R, Findeisen HM. 2018. Altered cellular metabolism drives trained immunity. Trends Endocrinol Metab 29:602–605. 10.1016/j.tem.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Erick TK, Brossay L. 2016. Phenotype and functions of conventional and non-conventional NK cells. Curr Opin Immunol 38:67–74. 10.1016/j.coi.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spits H, Bernink JH, Lanier L. 2016. NK cells and type 1 innate lymphoid cells: partners in host defense. Nat Immunol 17:758–764. 10.1038/ni.3482. [DOI] [PubMed] [Google Scholar]
- 12.Geiger TL, Sun JC. 2016. Development and maturation of natural killer cells. Curr Opin Immunol 39:82–89. 10.1016/j.coi.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahmoud AB, Tu MM, Wight A, Zein HS, Rahim MM, Lee SH, Sekhon HS, Brown EG, Makrigiannis AP. 2016. Influenza virus targets class I MHC-EDucated NK Cells for immunoevasion. PLoS Pathog 12:e1005446. 10.1371/journal.ppat.1005446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao H, Tu W, Qin G, Law HK, Sia SF, Chan PL, Liu Y, Lam KT, Zheng J, Peiris M, Lau YL. 2009. Influenza virus directly infects human natural killer cells and induces cell apoptosis. J Virol 83:9215–9222. 10.1128/JVI.00805-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strauss-Albee DM, Blish CA. 2016. Human NK cell diversity in viral infection: ramifications of ramification. Front Immunol 7:66. 10.3389/fimmu.2016.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao H, Tu W, Liu Y, Qin G, Zheng J, Chan PL, Lam KT, Peiris JS, Lau YL. 2010. Inhibition of human natural killer cell activity by influenza virions and hemagglutinin. J Virol 84:4148–4157. 10.1128/JVI.02340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. 2001. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 409:1055–1060. 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 18.Bar-On Y, Seidel E, Tsukerman P, Mandelboim M, Mandelboim O. 2014. Influenza virus uses its neuraminidase protein to evade the recognition of two activating NK cell receptors. J Infect Dis 210:410–418. 10.1093/infdis/jiu094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bar-On Y, Glasner A, Meningher T, Achdout H, Gur C, Lankry D, Vitenshtein A, Meyers AFA, Mandelboim M, Mandelboim O. 2013. Neuraminidase-mediated, NKp46-dependent immune-evasion mechanism of influenza viruses. Cell Rep 3:1044–1050. 10.1016/j.celrep.2013.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deniz G, Erten G, Kucuksezer UC, Kocacik D, Karagiannidis C, Aktas E, Akdis CA, Akdis M. 2008. Regulatory NK cells suppress antigen-specific T cell responses. J Immunol 180:850–857. 10.4049/jimmunol.180.2.850. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Zheng J, Liu Y, Wen L, Huang L, Xiang Z, Lam KT, Lv A, Mao H, Lau YL, Tu W. 2018. Uncompromised NK cell activation is essential for virus-specific CTL activity during acute influenza virus infection. Cell Mol Immunol 15:827–837. 10.1038/cmi.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcenaro E, Carlomagno S, Pesce S, Moretta A, Sivori S. 2011. Bridging innate NK cell functions with adaptive immunity. Adv Exp Med Biol 780:45–55. 10.1007/978-1-4419-5632-3_5. [DOI] [PubMed] [Google Scholar]
- 23.Schuster IS, Coudert JD, Andoniou CE, Degli-Esposti MA. 2016. “Natural Regulators”: NK cells as modulators of T cell immunity. Front Immunol 7:235. 10.3389/fimmu.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillespie AL, Teoh J, Lee H, Prince J, Stadnisky MD, Anderson M, Nash W, Rival C, Wei H, Gamache A, Farber CR, Tung K, Brown MG. 2016. Genomic modifiers of natural killer cells, immune responsiveness and lymphoid tissue remodeling together increase host resistance to viral infection. PLoS Pathog 12:e1005419. 10.1371/journal.ppat.1005419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heath J, Newhook N, Comeau E, Gallant M, Fudge N, Grant M. 2016. NKG2C+ CD57+ natural killer cell expansion parallels cytomegalovirus-specific CD8+ T cell evolution towards senescence. J Immunol Res 2016:7470124. 10.1155/2016/7470124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kos FJ, Engleman EG. 1995. Requirement for natural killer cells in the induction of cytotoxic T cells. J Immunol 155:578–584. [PubMed] [Google Scholar]
- 27.Loyon R, Picard E, Mauvais O, Queiroz L, Mougey V, Pallandre JR, Galaine J, Mercier-Letondal P, Kellerman G, Chaput N, Wijdenes J, Adotevi O, Ferrand C, Romero P, Godet Y, Borg C. 2016. IL-21-induced MHC class II+ NK cells promote the expansion of human uncommitted CD4+ central memory t cells in a macrophage migration inhibitory factor-dependent manner. J Immunol 197:85–96. 10.4049/jimmunol.1501147. [DOI] [PubMed] [Google Scholar]
- 28.Ge MQ, Ho AW, Tang Y, Wong KH, Chua BY, Gasser S, Kemeny DM. 2012. NK cells regulate CD8+ T cell priming and dendritic cell migration during influenza A infection by IFN-gamma and perforin-dependent mechanisms. J Immunol 189:2099–2109. 10.4049/jimmunol.1103474. [DOI] [PubMed] [Google Scholar]
- 29.Hoegl S, Ehrentraut H, Brodsky KS, Victorino F, Golden-Mason L, Eltzschig HK, McNamee EN. 2017. NK cells regulate CXCR2+ neutrophil recruitment during acute lung injury. J Leukoc Biol 101:471–480. 10.1189/jlb.3A0516-227R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capuano C, Battella S, Pighi C, Franchitti L, Turriziani O, Morrone S, Santoni A, Galandrini R, Palmieri G. 2018. Tumor-targeting anti-CD20 antibodies mediate in vitro expansion of memory natural killer cells: impact of CD16 affinity ligation conditions and in vivo priming. Front Immunol 9:1031. 10.3389/fimmu.2018.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, Heinrich F, Gasparoni G, Babic M, Tomic A, Pietra G, Nienen M, Blau IW, Hofmann J, Na IK, Prinz I, Koenecke C, Hemmati P, Babel N, Arnold R, Walter J, Thurley K, Mashreghi MF, Messerle M, Romagnani C. 2018. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol 19:453–463. 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Botet M, Muntasell A, Vilches C. 2014. The CD94/NKG2C+ NK-cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection. Semin Immunol 26:145–151. 10.1016/j.smim.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Lifshitz L, Gellatly K, Vinton CL, Busman-Sahay K, McCauley S, Vangala P, Kim K, Derr A, Jaiswal S, Kucukural A, McDonel P, Hunt PW, Greenough T, Houghton J, Somsouk M, Estes JD, Brenchley JM, Garber M, Deeks SG, Luban J. 2020. HIV-1-induced cytokines deplete homeostatic innate lymphoid cells and expand TCF7-dependent memory NK cells. Nat Immunol 21:274–286. 10.1038/s41590-020-0593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suliman S, Geldenhuys H, Johnson JL, Hughes JE, Smit E, Murphy M, Toefy A, Lerumo L, Hopley C, Pienaar B, Chheng P, Nemes E, Hoft DF, Hanekom WA, Boom WH, Hatherill M, Scriba TJ. 2016. Bacillus Calmette-Guerin (BCG) revaccination of adults with latent Mycobacterium tuberculosis infection induces long-lived BCG-reactive NK cell responses. J Immunol 197:1100–1110. 10.4049/jimmunol.1501996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerwenka A, Lanier LL. 2016. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol 16:112–123. 10.1038/nri.2015.9. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Wang J, Wang Y, Chen Y, Wei H, Sun R, Tian Z. 2017. Respiratory influenza virus infection induces memory-like liver NK cells in mice. J Immunol 198:1242–1252. 10.4049/jimmunol.1502186. [DOI] [PubMed] [Google Scholar]
- 37.Min-Oo G, Lanier LL. 2014. Cytomegalovirus generates long-lived antigen-specific NK cells with diminished bystander activation to heterologous infection. J Exp Med 211:2669–2680. 10.1084/jem.20141172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nabekura T, Lanier LL. 2016. Activating receptors for self-MHC class I enhance effector functions and memory differentiation of NK cells during mouse cytomegalovirus infection. Immunity 45:74–82. 10.1016/j.immuni.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH. 2010. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol 11:1127–1135. 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes TD, Bryceson YT. 2016. Natural killer cell memory in context. Semin Immunol 28:368–376. 10.1016/j.smim.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 41.O'Sullivan TE, Sun JC, Lanier LL. 2015. Natural killer cell memory. Immunity 43:634–645. 10.1016/j.immuni.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Boorn JG, Jakobs C, Hagen C, Renn M, Luiten RM, Melief CJ, Tuting T, Garbi N, Hartmann G, Hornung V. 2016. Inflammasome-dependent induction of adaptive NK cell memory. Immunity 44:1406–1421. 10.1016/j.immuni.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Wu LS, Wang JY. 2018. Warm up, cool down, and tearing apart in NK cell memory. Cell Mol Immunol 15:1095–1097. 10.1038/s41423-018-0188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikzad R, Angelo LS, Aviles-Padilla K, Le DT, Singh VK, Bimler L, Vukmanovic-Stejic M, Vendrame E, Ranganath T, Simpson L, Haigwood NL, Blish CA, Akbar AN, Paust S. 2019. Human natural killer cells mediate adaptive immunity to viral antigens. Sci Immunol 4:eaat8116. 10.1126/sciimmunol.aat8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dou Y, Fu B, Sun R, Li W, Hu W, Tian Z, Wei H. 2015. Influenza vaccine induces intracellular immune memory of human NK cells. PLoS One 10:e0121258. 10.1371/journal.pone.0121258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodier MR, Rodriguez-Galan A, Lusa C, Nielsen CM, Darboe A, Moldoveanu AL, White MJ, Behrens R, Riley EM. 2016. Influenza vaccination generates cytokine-induced memory-like NK Cells: impact of human cytomegalovirus infection. J Immunol 197:313–325. 10.4049/jimmunol.1502049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou K, Wang J, Li A, Zhao W, Wang D, Zhang W, Yan J, Gao GF, Liu W, Fang M. 2016. Swift and strong NK cell responses protect 129 mice against high-dose influenza virus infection. J Immunol 196:1842–1854. 10.4049/jimmunol.1501486. [DOI] [PubMed] [Google Scholar]
- 48.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. 2009. Maturation of mouse NK cells is a 4-stage developmental program. Blood 113:5488–5496. 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 49.Liu WC, Lin YL, Spearman M, Cheng PY, Butler M, Wu SC. 2016. Influenza hemagglutinin glycoproteins with different N-glycan patterns activate dendritic cells in vitro. J Virol 90:6085–6096. 10.1128/JVI.00452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zylicz M, King FW, Wawrzynow A. 2001. Hsp70 interactions with the p53 tumour suppressor protein. EMBO J 20:4634–4638. 10.1093/emboj/20.17.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milo I, Blecher-Gonen R, Barnett-Itzhaki Z, Bar-Ziv R, Tal O, Gurevich I, Feferman T, Drexler I, Amit I, Bousso P, Shakhar G. 2018. The bone marrow is patrolled by NK cells that are primed and expand in response to systemic viral activation. Eur J Immunol 48:1137–1152. 10.1002/eji.201747378. [DOI] [PubMed] [Google Scholar]
- 52.Kvell K, Cooper EL, Engelmann P, Bovari J, Nemeth P. 2007. Blurring borders: innate immunity with adaptive features. Clin Dev Immunol 2007:83671. 10.1155/2007/83671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrillo-Bustamante P, Keşmir C, de Boer RJ. 2016. The evolution of natural killer cell receptors. Immunogenetics 68:3–18. 10.1007/s00251-015-0869-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahim MM, Makrigiannis AP. 2015. Ly49 receptors: evolution, genetic diversity, and impact on immunity. Immunol Rev 267:137–147. 10.1111/imr.12318. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Beltran WF, Holzemer A, Martrus G, Chung AW, Pacheco Y, Simoneau CR, Rucevic M, Lamothe-Molina PA, Pertel T, Kim TE, Dugan H, Alter G, Dechanet-Merville J, Jost S, Carrington M, Altfeld M. 2016. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat Immunol 17:1067–1074. 10.1038/ni.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walter L, Petersen B. 2017. Diversification of both KIR and NKG2 NK cell receptor genes in macaques - implications for highly complex MHC-dependent regulation of NK cells. Immunology 150:139–145. 10.1111/imm.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duev-Cohen A, Bar-On Y, Glasner A, Berhani O, Ophir Y, Levi-Schaffer F, Mandelboim M, Mandelboim O. 2016. The human 2B4 and NTB-A receptors bind the influenza viral hemagglutinin and co-stimulate NK cell cytotoxicity. Oncotarget 7:13093–13105. 10.18632/oncotarget.7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, Riley JK, Zhu J, Tian Z, Yokoyama WM. 2014. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife 3:e01659. 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bauer M, Weis S, Netea MG, Wetzker R. 2018. Remembering pathogen dose: long-term adaptation in innate immunity. Trends Immunol 39:438–445. 10.1016/j.it.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Kugelberg E. 2016. Immune memory: inflammasomes drive NK cell memory. Nat Rev Immunol 16:405–405. 10.1038/nri.2016.74. [DOI] [PubMed] [Google Scholar]
- 61.Kronstad LM, Seiler C, Vergara R, Holmes SP, Blish CA. 2018. Differential induction of IFN-alpha and modulation of CD112 and CD54 expression govern the magnitude of NK cell IFN-gamma response to influenza A viruses. J Immunol 201:2117–2131. 10.4049/jimmunol.1800161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geary CD, Sun JC. 2017. Memory responses of natural killer cells. Semin Immunol 31:11–19. 10.1016/j.smim.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ram DR, Manickam C, Lucar O, Shah SV, Reeves RK. 2019. Adaptive NK cell responses in HIV/SIV infections: a roadmap to cell-based therapeutics? J Leukoc Biol 105:1253–1259. 10.1002/JLB.MR0718-303R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberto A, Di Vito C, Zaghi E, Mazza EMC, Capucetti A, Calvi M, Tentorio P, Zanon V, Sarina B, Mariotti J, Bramanti S, Tenedini E, Tagliafico E, Bicciato S, Santoro A, Roederer M, Marcenaro E, Castagna L, Lugli E, Mavilio D. 2018. The early expansion of anergic NKG2Apos/CD56dim/CD16neg natural killer cells represents a therapeutic target in haploidentical haematopoietic stem cell transplantation. Haematologica 103:1390–1402. 10.3324/haematol.2017.186619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, Blery M, Bonnafous C, Gauthier L, Morel A, Rossi B, Remark R, Breso V, Bonnet E, Habif G, Guia S, Lalanne AI, Hoffmann C, Lantz O, Fayette J, Boyer-Chammard A, Zerbib R, Dodion P, Ghadially H, Jure-Kunkel M, Morel Y, Herbst R, Narni-Mancinelli E, Cohen RB, Vivier E. 2018. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell 175:1731.e13–1743.e13. 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gulic T, Laskarin G, Dominovic M, Glavan Gacanin L, Babarovic E, Haller H, Rukavina D. 2016. Potential role of heat-shock protein 70 and interleukin-15 in the pathogenesis of threatened spontaneous abortions. Am J Reprod Immunol 76:126–136. 10.1111/aji.12525. [DOI] [PubMed] [Google Scholar]
- 67.Hwang CY, Holl J, Rajan D, Lee Y, Kim S, Um M, Kwon KS, Song B. 2010. Hsp70 interacts with the retroviral restriction factor TRIM5alpha and assists the folding of TRIM5alpha. J Biol Chem 285:7827–7837. 10.1074/jbc.M109.040618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aloni-Grinstein R, Charni-Natan M, Solomon H, Rotter V. 2018. p53 and the viral connection: back into the future‡. Cancers (Basel) 10:178. 10.3390/cancers10060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng J, Liu Y, Liu Y, Liu M, Xiang Z, Lam KT, Lewis DB, Lau YL, Tu W. 2013. Human CD8+ regulatory T cells inhibit GVHD and preserve general immunity in humanized mice. Sci Transl Med 5:168ra9. 10.1126/scitranslmed.3004943. [DOI] [PubMed] [Google Scholar]
- 71.Zheng J, Wu WL, Liu Y, Xiang Z, Liu M, Chan KH, Lau SY, Lam KT, To KK, Chan JF, Li L, Chen H, Lau YL, Yuen KY, Tu W. 2015. The therapeutic effect of pamidronate on lethal avian influenza A H7N9 virus infected humanized mice. PLoS One 10:e0135999. 10.1371/journal.pone.0135999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data files have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE124321.