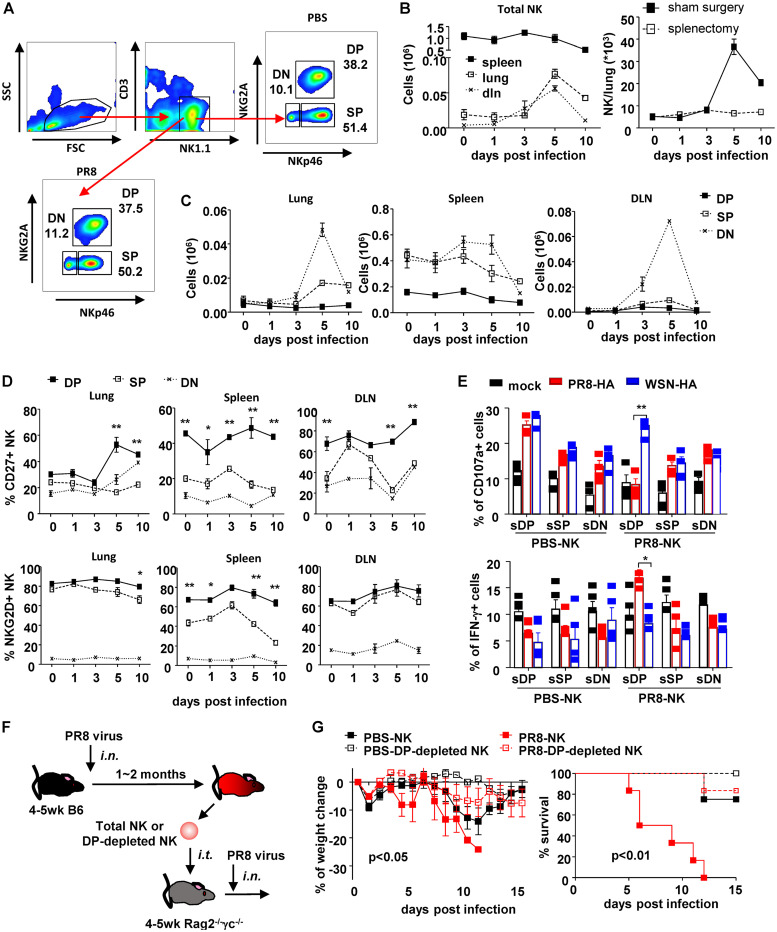
FIG 3.
NKp46+ NKG2A+ identified memory NK cell subset. (A) Flow chart of NK cell subsets identified by NKp46 and NKG2A in mock-treated mice and mice recovering from PR8 infection. DN, NKp46− NKG2A−; SP, NKp46+ NKG2A−; DP, NKp46+ NKG2A+. Dynamic changes of total NK cells (B, left) and NK subsets (C) in lung, DLN, and spleen of mice infected with sublethal dose of PR8 virus (25 μl, 103.5 TCID50), i.n., were determined by flow cytometry with counting beads. One month postinfection, some completely recovered mice received splenectomy or sham surgery. One month postsurgery, mice were infected with lethal dose of PR8 virus (25 μl, 105 TCID50), i.n. Lung-infiltrating NK cells were counted by flow cytometry at indicated times postinfection (B, right). (D) Expressions of CD27 and NKG2D in NK cell subsets in lung, DLN, and spleen during infection were determined by surface staining and flow cytometry. Difference between SP and DP populations were statistically analyzed. (E) Expressions of CD107a and IFN-γ in flow cytometry-separated NK cell subsets (sDP, sSP, and sDN) (effector [E]) were determined by surface and intracellular staining and flow cytometry after cocultured with recombinant virus-treated DCs (target [T]) at an E/T ratio of 10:1 for 4 h at 37°C and 5% CO2; n = 4. (F) Protocol of NK cells subset isolation and adoptive transfer. (G) Weight change and survival of recipient mice. DP, NKp46+ NKG2A+. Data are shown as means ± SEMs and represent 3 independent experiments; n = 4 to 6. *, P < 0.05; **, P < 0.01.