FIG 3.
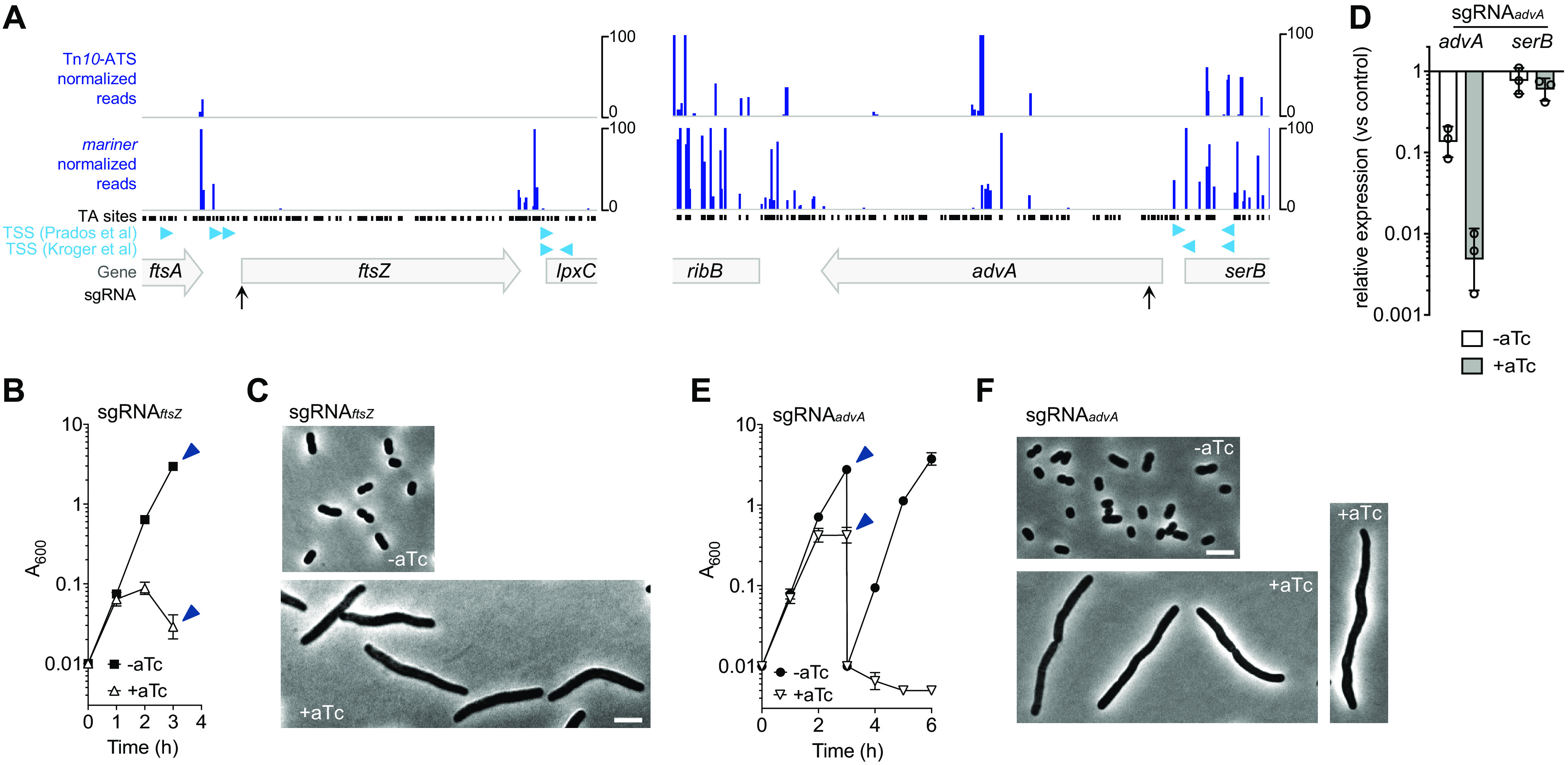
CRISPRi knockdown of ftsZ and advA blocks growth and cell division. (A) Transposon mutations in ftsZ and in most regions of advA were not detectable within Tn-seq pools. Normalized Tn-seq read counts were plotted according to transposon insertion position in mariner and Tn10-ATS libraries. Black points show locations of potential mariner insertion sites (TA dinucleotides). Predicted TSSs are as described in references 24 (Kroger et al.) and 28 (Prados et al.). Positions targeted by sgRNAs are indicated as in Fig. 2. (B) YDA004 with sgRNAftsZ was cultured with or without 200 ng/ml aTc, and growth was monitored. Data points show geometric mean A600 ± SD (n = 3). At 3 h, samples were collected (arrowheads) and imaged by phase-contrast microscopy (C). (D) Efficient and specific CRISPRi knockdown of advA. YDA004 cells with sgRNAadvA or the nontargeting control were cultured for 2 h with aTc (200 ng/ml) or no inducer, and advA and serB transcription levels relative to that in the untreated control were analyzed via qRT-PCR. Bars show geometric mean ± SD (n = 3). In unpaired t tests comparing sgRNAadvA with the control, advA transcription showed a significant decrease with (P = 0.008) or without (P = 0.01) aTc; serB transcription showed no significant change (P > 0.07). (E and F) AdvA is essential in LB. Growth (E) and morphology (from samples indicated by arrowheads [F]) were analyzed as in panels B and C. Data points show geometric mean A600 ± SD (n = 3). Scale bars = 5 μm.
