Figure 2.
Detection of endogenous L1-mediated retrotransposition in human cells
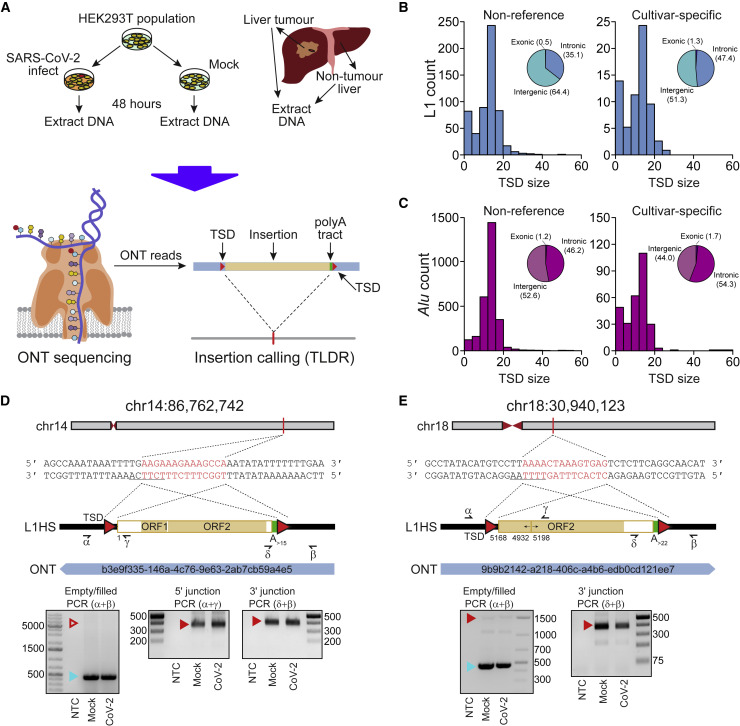
(A) Experimental design. HEK293T cells were divided into two populations (cultivars), which were then either SARS-CoV-2 infected or mock infected. DNA was extracted from each cultivar, as well as from hepatocellular carcinoma patient samples, and subjected to ONT sequencing. ONT reads were used to call non-reference L1 and virus insertions with TLDR, which also resolves TSDs and other retrotransposition hallmarks. TSDs, red triangles; polyA tract, green rectangle; ONT read, blue rectangle. Some illustrations are adapted from a previous study (Ewing et al., 2020).
(B) TSD size distribution for non-reference L1 insertions, as annotated by TLDR, as well as cultivar-specific L1 insertions found only in either our HEK293T cells infected with SARS-CoV-2 or our mock infected cells. Pie charts indicate the percentages of exonic, intronic, and intergenic insertions, annotated by RefSeq coordinates.
(C) As for (B), except showing data for Alu insertions.
(D) Detailed characterization of an L1 insertion detected in SARS-CoV-2-infected HEK293T cells by a single spanning ONT read aligned to chromosome 14. Nucleotides highlighted in red correspond to the integration site TSD. Underlined nucleotides correspond to the L1 EN motif. The cartoon indicates a full-length L1HS insertion flanked by TSDs (red triangles) and a 3ʹ polyA tract (green), with the underneath numeral representing the 5ʹ-end position relative to the mobile L1HS sequence L1.3 (Dombroski et al., 1993). The relevant spanning ONT read, with identifier, is also positioned underneath the cartoon. Symbols (α, β, δ, and γ) represent the approximate position of primers used for empty/filled site and L1-genome junction PCR validation reactions. They are displayed in gel images if successful. Ladder band sizes are as indicated; NTC, non-template control. Red triangles indicate L1 amplicon expected sizes (empty triangle, no product; filled triangle, capillary sequenced on-target product). Blue triangles indicate expected empty site sizes.
(E) As for (D), except for a 5ʹ inverted/deleted L1HS located on chromosome 18.
Please see Figures S1 and S2 and Tables S1 and S2 for further information.