Figure 3.
ONT reads occasionally align to viral genome sequences
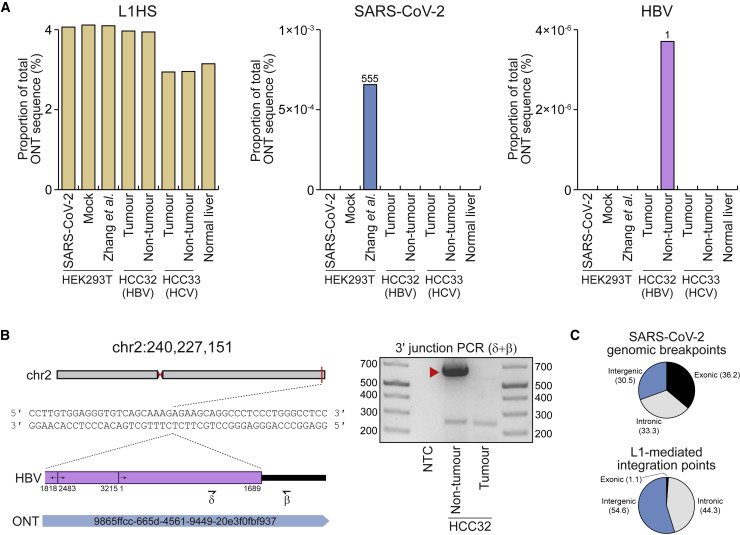
(A) Percentages of total ONT sequence alignable to L1HS (left), SARS-CoV-2 (middle), and HBV (right) isolate genomes. Read counts for SARS-CoV-2 and HBV are provided above histogram columns. No reads were aligned to the HCV isolate genomes. HEK293T data were generated here (SARS-CoV-2, mock) or by Zhang et al.. HCC tumor/non-tumor liver pairs were sequenced here (HCC32; confirmed HBV-positive) or previously (Ewing et al., 2020) (HCC33; HCV-positive). Normal liver ONT sequencing from our prior work (Ewing et al., 2020) was included as an additional control.
(B) A HBV insertion detected in non-tumor liver. In this example, an ONT read from the non-tumor liver of HCC32 spanned the 3ʹ junction of a HBV integrant located on chromosome 2. Of the HBV isolate genomes considered here, this read aligned best to a representative of genotype B (GenBank: AB602818). The HBV sequence was rearranged consistent with its linearization prior to integration (Fujimoto et al., 2012; Jiang et al., 2012; Nagaya et al., 1987). Numerals indicate positions relative to AB602818. Symbols (β and δ) represent the approximate position of primers used to PCR validate the HBV insertion. The gel image at right shows the 3ʹ junction PCR results. Ladder band sizes are as indicated. The red-filled triangle indicates an on-target product confirmed by capillary sequencing. Repeated attempts to PCR amplify the 5ʹ junction of the HBV integrant did not return an on-target product, perhaps due to a genomic deletion at the insertion site.
(C) Percentages of exonic (black), intronic (gray), and intergenic (blue) genomic alignment breakpoints for ONT reads also aligned to the SARS-CoV-2 genome and also for the non-reference L1-mediated insertions reported here. Genomic features were annotated according to RefSeq coordinates.