FIG 3.
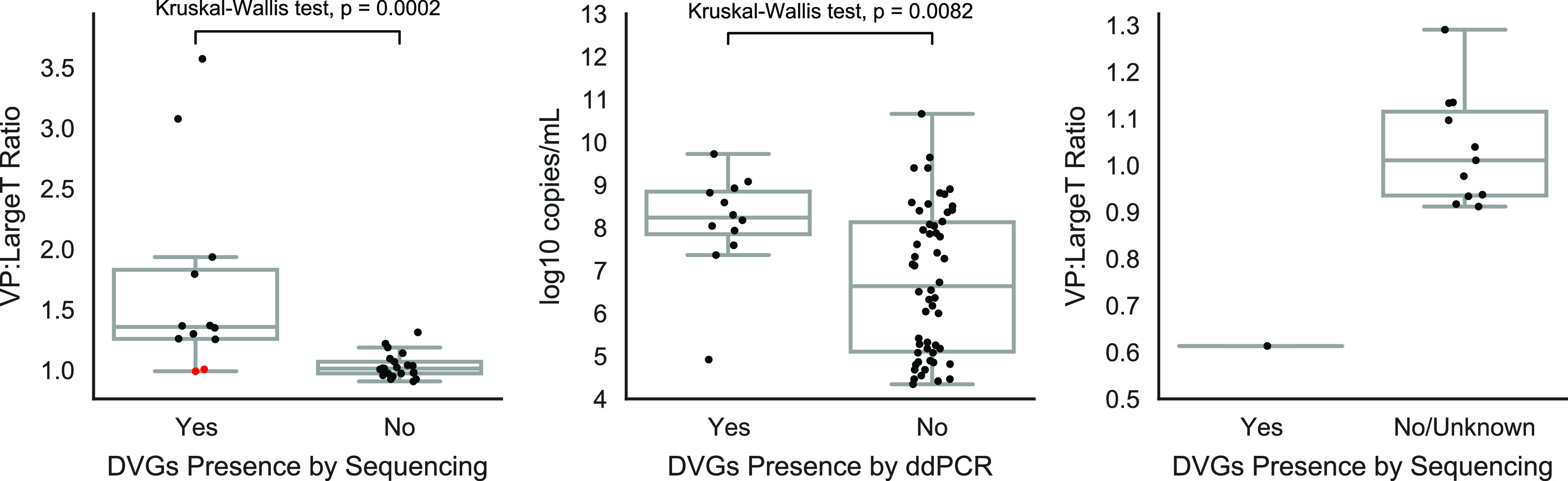
Confirmation of defective viral genomes using droplet digital PCR. (A) The copy number ratio for VP1 and large T antigen is plotted for 33 BKPyV isolates with and without DVGs determined by sequencing. Quartiles for each group are plotted in a box-and-whiskers plot, and error bars are 1.5-fold the interquartile range. Red dots indicate two strains (UBK265/UBK266) with DVGs that had a comparably low level of rearrangements (3 to 5%) at the target loci as determined by sequencing, consistent with their equivalent copy number measured by ddPCR. Statistical comparison was performed via Kruskal-Wallis test. (B) Log10 copies/ml viral load values in 66 BKPyV-positive specimens are displayed. The presence of DVGs was determined by ddPCR using a VP1/large T antigen ratio cutoff of 1.25. Statistical comparison was performed via Kruskal-Wallis test. (C) ddPCR VP1/large T antigen copy number ratios for 12 JCPyV-positive specimens are depicted. One JCPyV specimen contained a sequence-confirmed DVG, while 7 of the 11 other specimens lacked DVGs by sequencing, and the remaining had unknown DVG status.
