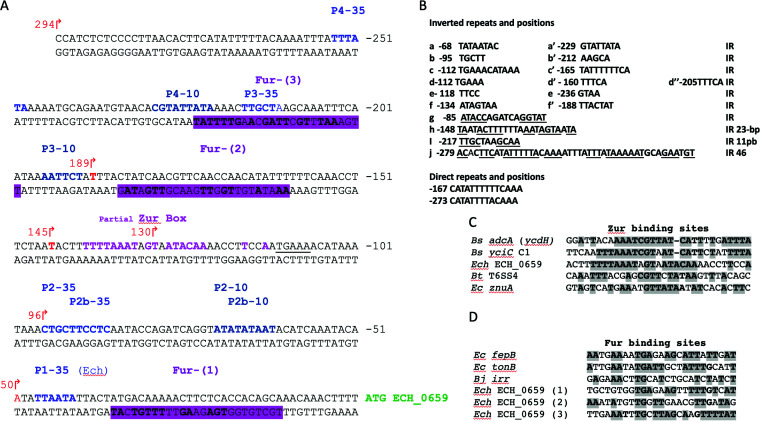
FIG 1.
The putative promoter region upstream of the ECH_0659 coding region contains DNA binding motifs that likely contribute to metal ion regulation and transcription. (A) Putative promoter region upstream of the ECH_0659 coding region. Regions that resemble the consensus for the Fur-binding site are indicated by magenta shading and in boldface type and are labeled Fur-(1), Fur-(2), and Fur-(3). The region that resembles the consensus for the Zur-binding site is indicated in boldface type in magenta color and labeled “partial Zur box.” Predicted −35 and −10 sequences are shown in blue and cyan-blue text, respectively, and are labeled P1-35, P2-35, P2b-35, P3-35, P4-35, P2-10, P2b-10, P3-10, and P4-10. (B) Inverted repeat sequences that are separated are a-a′, b, b′, c-c′, d-d′-d″, e-e′ and f-f′; inverted repeats that are located at close proximity are identified with underlines and listed as h, i, and j. Direct repeats are shown after the inverted repeats. (C and D) Zur (C) and Fur (D) partial consensus binding site sequences from other Gram-positive and Gram-negative bacteria are indicated by gray shading. Bt, Burkholderia thailandensis; Bs, Bacillus subtilis; Bj, Bradyrhizobium japonicum; Ec, E. coli; Ech, E. chaffeensis.