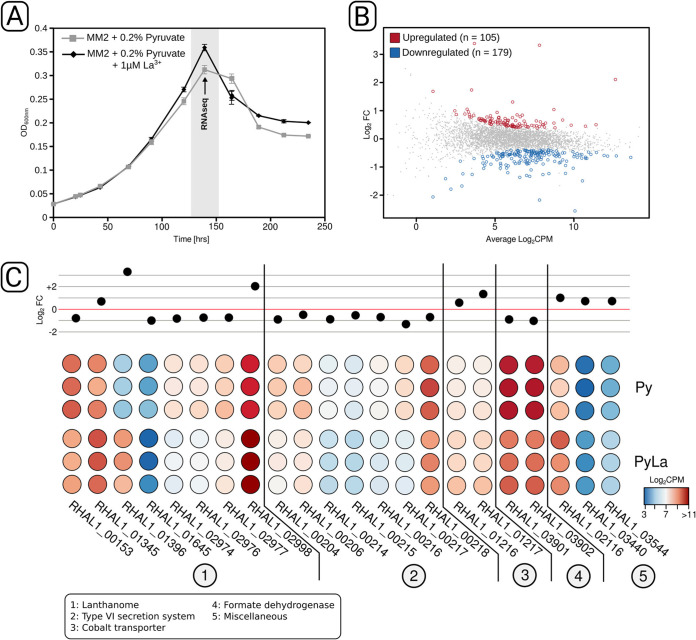
FIG 6.
RNA-seq analysis of Beijerinckiaceae bacterium RH AL1 grown with and without lanthanum under heterotrophic conditions. Pyruvate was used as a carbon source for cultivation in the MM2 medium. (A) Lanthanum supplementation led to higher maximum optical densities. OD600nm, optical density at 600 nm. (B) Biomass samples after 6 days (144 h, marked in gray) of incubation were harvested and used for RNA-seq library preparation. RNA-seq-based, differential gene expression analysis revealed 284 differentially expressed genes. (C) Fold changes and counts per million are shown for selected differentially expressed genes. The red line in the upper half represents no fold change. The color code of the heatmap indicates counts per million. Genes are grouped according to the functions of proteins for which they code. A list with all differentially expressed genes and their annotation can be found in Table S4 in the supplemental material. Log2FC, log2 fold change, Log2CPM, log2 counts per million.