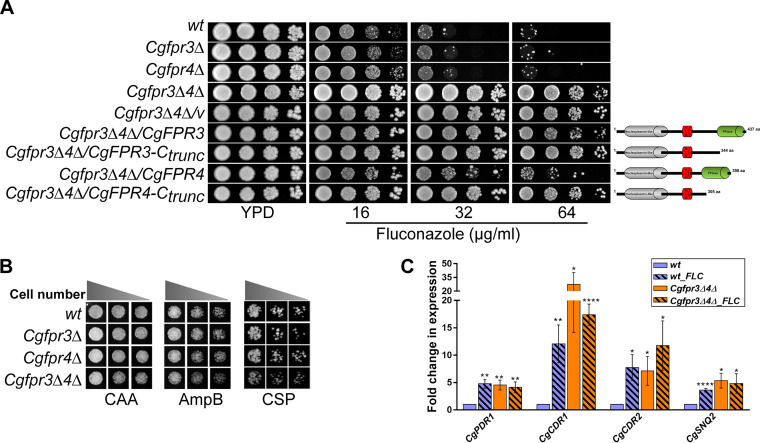
FIG 3.
The Cgfpr3Δ4Δ mutant is resistant to fluconazole. (A) Serial dilution spotting analysis indicating the fluconazole susceptibility of the indicated C. glabrata strains. The domain compositions of full-length and C-terminally truncated CgFpr3 and CgFpr4 proteins are shown schematically on the right side of the spotting image. NLS, nuclear localization signal. (B) Liquid medium-based growth analysis of the indicated C. glabrata strains to assess sensitivity to amphotericin B and caspofungin. wt and CgfprΔ mutant strains were grown in Casamino acids medium lacking (CAA) or containing amphotericin B (AmpB) (500 ng/ml) and caspofungin (CSP) (125 ng/ml) at 30°C. After 24 h of incubation, cultures were diluted in PBS, and 3 μl of 100-, 250-, and 500-fold-diluted cultures was spotted onto YPD medium. Images were captured after 1 day of growth at 30°C. (C) qPCR-based quantification of CgCDR1, CgCDR2, CgPDR1, and CgSNQ2 transcript levels. Log-phase wt and Cgfpr3Δ4Δ cells were either treated with 16 μg/ml fluconazole for 4 h or left untreated. RNA was extracted, and qPCR was set up using 500 ng of total RNA. Transcript levels were quantified using the 2−ΔΔCT method. Data (means ± SEM; n = 3 to 5) were normalized against the CgTDH3 mRNA as a control and represent fold changes in the expression of the CgCDR1, CgCDR2, CgPDR1, and CgSNQ2 genes in untreated Cgfpr3Δ4Δ and fluconazole (FLC)-treated wt and Cgfpr3Δ4Δ cells, compared to the untreated wt cells (taken as 1.0). *, P ≤ 0.0332; **, P ≤ 0.0021; ****, P ≤ 0.0001 (by multiple t tests).