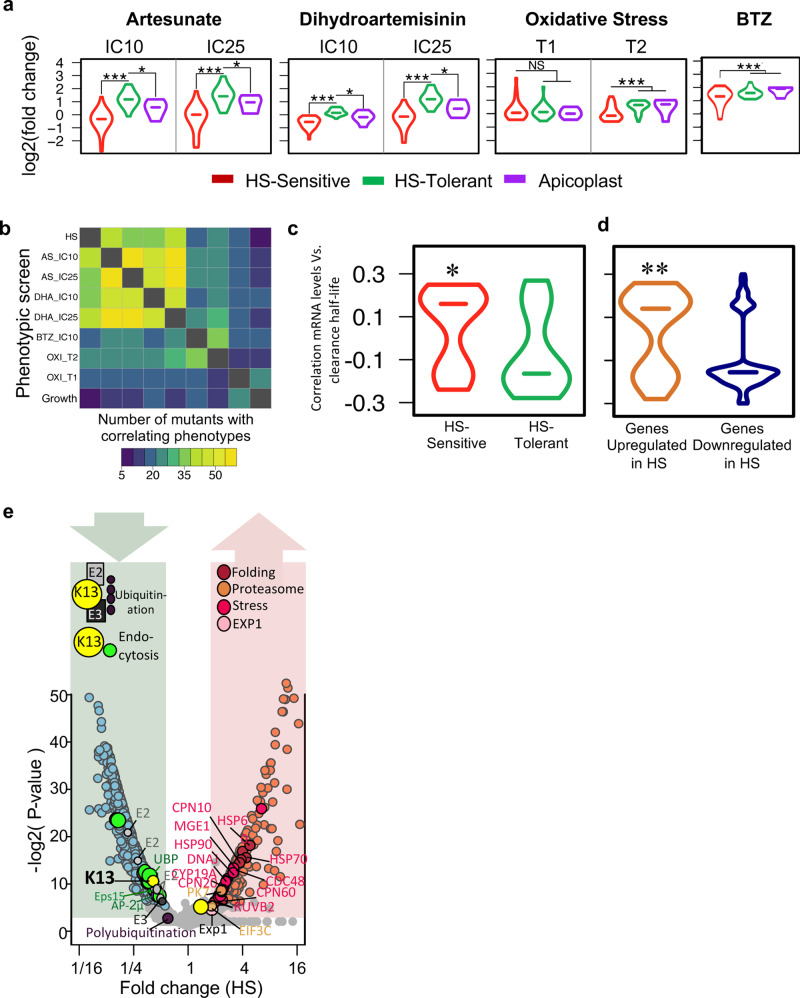
Fig. 6. Increased sensitivity to fever is directly correlated with increased sensitivity to artemisinin in the malaria parasite.
a HS-Sensitive pB-mutants (red) are more sensitive to artemisinin derivatives AS and DHA, proteasome-inhibitor BTZ, and conditions of heightened oxidative stress (T1 and T2, “Methods” section) than HS-tolerant parasites (green) in all pooled screens of the pilot library. HS-Sensitive mutants tended to be sensitive to both artemisinin derivatives and H2O2-induced oxidative stress, while HS-Tolerant mutants were less sensitive to either condition. HS-Sensitive mutants also had increased sensitivity to BTZ (Supplementary Data File 4 and Supplementary Figs. 2, 10). Biological replicates (n = 2) were highly correlated for all screens (Pearson correlation >0.94; Supplementary Figs. 2 and 10). Mutants in apicoplast-targeted genes (purple; n = 5) and HS-Sensitive mutants (n = 28) had similar phenotypes to artemisinin derivatives, but not to proteasome inhibitors or oxidative stress (*two-tailed Wilcoxon p < 0.05; ***two-tailed Wilcoxon p < 1e−10). b Correlation between mutant phenotypes in all pooled screens of the pilot library. Mutants performing in the bottom 25% or top 25% of each screen were classified as Sensitive and Tolerant, respectively. Mutants falling into the same category in pairwise comparisons between screens were considered to have correlating phenotypes. c Compared with HS-Tolerant genes (green, n = 16 mutants), mRNA levels of HS-Sensitive genes (red, n = 29 mutants) are positively correlated with parasite clearance half-life under ACT pressure in field isolates15. (*two-tailed Wilcoxon p-value < 0.05). d Genes upregulated in HS (red, n = 67) are positively correlated with parasite clearance half-life under ACT in field isolates15. Downregulated genes are more likely to be negatively correlated with parasite clearance half-life (green, n = 114). (**two-tailed Wilcoxon p-value < 1e−3). e Both K13-mediated mechanisms of artemisinin resistance (endocytosis, ubiquitin-proteasome system) are similarly regulated in HS. RNAseq data are plotted by average log2 fold-change in HS and significance (−log2 of FDR-corrected two-tailed Fisher test). Circles in shades of blue and pink indicate genes downregulated or upregulated in HS, respectively. Source data are provided as a Source Data file.